Abstract
Portulaca oleracea is an important and widely distributed medicinal and edible plant, which has great economic value in the medical and food industries in the whole world. The complete chloroplast genome of Portulaca oleracea was found to possess a total length 156,533 bp and had the circular structure. It was the typical quadripartite structure the same as many plants, including 87,437 bp large single-copy region (LSC), 18,096 bp small single-copy region (SSC) of and a pair of 25,500 bp inverted repeat (IR) regions. The total of 132 genes was successfully annotated, which contained 87 protein-coding genes, 37 transfer RNA genes, and eight ribosomal RNA genes. Nineteen genes were found in one of IR, which included eight protein-coding gene, seven tRNA genes, and four rRNA genes. The overall nucleotide composition of chloroplast genome sequence is: 31.5% (A), 32.1% (T), 18.5% (C), 17.9% (G), and the total GC content of 36.4%. The phylogenetic analysis result showed that Portulaca oleracea was close to Carnegiea gigantean in the phylogenetic relationship by the neighbor-joining (NJ) method in this study.
Portulaca oleracea is listed by the World Health Organization (WHO) as one of the most used medicinal and edible plants, which is packed full of nutrients, vitamins, and minerals. Portulaca oleracea has been widely used as a potherb and herb in the Mediterranean, central European, and Asian countries (Iranshahy et al. Citation2017). Portulaca oleracea has been used as a traditional Chinese medicine (TCM) in China, acting as a febrifuge, antiseptic, vermifuge, and so forth (Rahimi et al. Citation2019). It is considered as a beneficial herb in the world and contains antimicrobial, antidiabetic, and anti-inflammatory properties (Iranshahy et al. Citation2017). In this study, the complete chloroplast genome of Portulaca oleracea was characterized and generated, which can be used for the active ingredient of TCM and drug development in this species.
The fresh Portulaca oleracea were collected from the Hunan University of Chinese Medicine that located at Changsha, Hunan, and China (112.90E, 28.13 N), which were stored in the −80 °C refrigerator. Total genomic DNA of Portulaca oleracea was extracted from the fresh leaves with a modified CTAB method and sequenced, which was stored in Hunan University of Chinese Medicine (No.HNUCM-001). Here, the quality controlled and removed the sequences used by FastQC (Andrews Citation2015). The chloroplast genome of Portulaca oleracea was assembled by MitoZ (Meng et al. Citation2019) and annotated by geneious (Kearse et al. Citation2012). All the coding and other genes were predicted by CPGAVAS (Liu et al. Citation2012) and NCBI Blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi) in the chloroplast genome. Finally, the circular chloroplast genome map was generated by DOGMA (Wyman et al. Citation2004).
The complete chloroplast genome of Portulaca oleracea (NCBI No. NK9714231) was found to possess a total length 156,533 bp and had the circular structure. It was the typical quadripartite structure the same as many plants, including 87,437 bp large single-copy region (LSC), 18,096 bp small single-copy region (SSC) of and a pair of 25,500 bp inverted repeat (IR) regions. The total of 132 genes was successfully annotated, which contained 87 protein-coding genes, 37 transfer RNA genes, and 8 ribosomal RNA genes. Here, 19 genes were found in one of the IR, which included eight protein-coding genes (rps19, rpl2, rpl23, ycf2, ndhB, rps7, rps12, and ycf1), seven tRNA genes (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, and trnN-GUU) and four rRNA genes (rrn16, rrn23, rrn4.5, and rrn5). The overall nucleotide composition of chloroplast genome sequence is: 31.5% (A), 32.1% (T), 18.5% (C), 17.9% (G), and the total GC content of 36.4%, respectively.
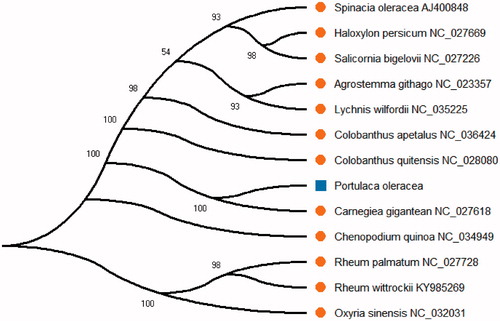
The neighbor-joining (NJ) tree was reconstructed using the complete chloroplast genome sequence of Portulaca oleracea with other published 12 species from NCBI (). All the sequences from NCBI were aligned by MAFFT (Katoh and Standley Citation2013) and the NJ phylogenetic analyses were conducted by MEGA X (Kumar et al. Citation2018). We used MEGA X with 2000 bootstraps values under the substitution model to reconstruct the NJ phylogenetic tree. At last, the NJ phylogenetic tree was drawn and edited using MEGA X (Kumar et al. Citation2018). The phylogenetic analysis result showed that Portulaca oleracea was close to Carnegiea gigantean in the phylogenetic relationship by the NJ method in this study. The result can be used for the active ingredient of TCM and drug development in this species.
Disclosure statement
No potential conflict of interest was reported by the author(s). This study does not contain any studies with human participants or animals performed by any of the authors.
Data availability statement
The data that support the findings of this study are openly available in Portulaca oleracea at http://doi.org/10.1080/23802359.2020.1791016, reference number [reference number NK9714231]. The data that support the findings of this study are available from the corresponding author, upon reasonable request.
References
- Andrews S. 2015. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Iranshahy M, Javadi B, Iranshahi M, Jahanbakhsh SP, Mahyari S, Hassani FV, Karimi G. 2017. A review of traditional uses, phytochemistry and pharmacology of Portulaca oleracea L. J Ethnopharmacol. 205:158–172.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715–2164.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Rahimi VB, Ajam F, Rakhshandeh H, Askari VR. 2019. A pharmacological review on Portulaca oleracea L.: focusing on anti-inflammatory, anti- oxidant, immuno-modulatory and antitumor activities. J Pharmacopuncture. 22(1):7–15.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.