Abstract
The complete mitochondrial genome of Linoclostis gonatias (Lepidoptera: Xyloryctidae) is 15,528 bp in length, containing 13 protein-coding genes, 22 transfer RNAs, 2 ribosomal RNAs, and a putative control region. Except for cox1 starts with CGA, all other PCGs use the typical ATN codons. Most of the PCGs end with the complete stop codon TAA, whereas cox2 terminates with the incomplete stop codon T. The BI analysis was performed using a dataset matrix containing 13 PCGs concatenated from the mitogenomes of Gelechioidea species. The monophyly of Xyloryctidae was highly supported. In addition, Oecophoridae was inferred as the sister group of Xyloryctidae.
The Xyloryctidae was often considered to be a subfamily of Oecophoridae, however, Hodges (Citation1999) raised it to family level. It is worldwide in distribution with more than 1200 species in 86 genera (Hodges, Citation1999). To date, only one complete mitochondrial genome (mitogenome) has been sequenced from members of this family. In this study, we present the complete mitogenome of Linoclostis gonatias (Lepidoptera: Xyloryctidae). It is reported as the major pest of Camellia oleifera, which is the most economically important woody oil tree in China.
The specimen was collected from Jiangxi Province, China (27.922227, 115.087795) in June 2019, and was preserved at Entomological Specimen Room of Jinggangshan University (accession number: 20190601S). Total genomic DNA was extracted from a single specimen and was sequenced by Illumina HiSeq2000, with a read length of 150 bp. The complete mitogenome was assembled by the GetOrganelle v1.6.4 program (Jin et al. Citation2018) and was annotated using the MITOS2 webserver (Bernt et al. Citation2013).
The complete mitogenome of L. gonatias is a circular molecule of 15,528 bp in length. It contains the typical set of 37 genes as in most insect mitogenomes (Cameron Citation2014), including 13 protein-coding genes (PCGs), 2 ribosomal RNAs (rrnL and rrnS), 22 transfer RNAs (tRNAs), and a non-coding control region. The J-strand encodes most of the genes (9 PCGs and 14 tRNAs), while the remaining genes (4 PCGs, 8 tRNAs, and 2 rRNAs) are located on the N-strand. The overall base composition is 39.3% A, 40.9% T, 12.2% C, and 7.5% G. The nucleotide composition of the L. gonatias mitogenome is biased toward A + T (80.2%). All of the PCGs use the standard start codon ATN, except for cox1 which starts with CGA. Twelve PCGs use the common stop codon TAA, however, cox2 terminates with the incomplete stop codon T. The rrnL is located between trn-Leu1 and trn-Val, with the length of 1345 bp. The rrnS is located between trn-Val and the control region, with the length of 791 bp. The control region is 313 bp in length, and is located between rrnS and trn-Ile. Like the majority of other lepidopterans (Wang et al., Citation2016), it is characterized by its remarkably high A + T content of 95.2%.
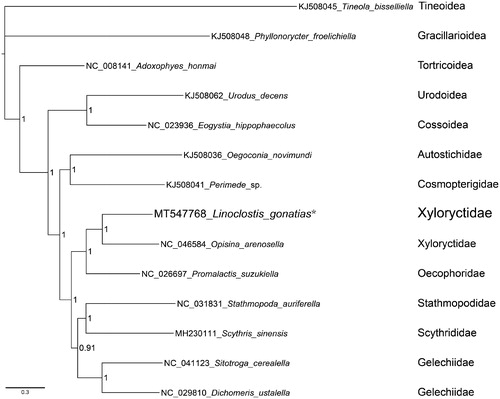
Phylogenetic analysis was performed on concatenated nucleotide sequences of the 13 PCGs of nine Gelechioidea species, along with five outgroup species (). The best-fit partitioning scheme and nucleotide substitution model were simultaneously confirmed with PartitionFinder 2.1.1 (Lanfear et al. Citation2016). Phylogenetic inference was performed using MrBayes 3.2.6 (Ronquist et al., Citation2012) through the online CIPRES Science gateway (Miller et al., Citation2010). Within the Gelechioidea, L. gonatias was grouped with Opisina arenosella as the sister group. Accordingly, the monophyly of Xyloryctidae was highly supported. In addition, Oecophoridae was inferred as the sister group of Xyloryctidae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available in [GenBank] at [https://www.ncbi.nlm.nih.gov/nuccore/MT547768], reference number [MT547768].
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117.
- Hodges RW. 1999. The Gelechioidea. Vol. IV, Part 35. In: Kristensen NP, edtior. Lepidoptera: moths and butterflies. 1. Evolution, systematics, and biogeography. Handbook of zoology. Berlin and New York: De Gruyter; p. 131–158.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. BioRxiv. 256479.DOI:10.1101/256479
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2016. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol E. 34:772–773.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE). Gateway Computing Environments Workshop (GCE). 14:1–8.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Wang QQ, Zhang ZQ, Tang GH. 2016. The mitochondrial genome of Atrijuglans hetaohei Yang (Lepidoptera: Gelechioidea) and related phylogenetic analyses. Gene. 581(1):66–74.