Abstract
Manglietia longirostrata Sima is a rare and endemic species in China. The complete chloroplast genome (cpDNA) of M. longirostrata was sequenced and assembled in this study. The cpDNA is 160,049 bps in length, contains a large single-copy region (LSC) of 88,098 bp and a small single-copy region (SSC) of 18,861 bp, separated by a pair of identical inverted repeat (IR) regions of 26,571 bp, each. The genome contains 123 genes, including 73 protein-coding genes, 8 ribosomal RNA genes, and 37 transfer RNA genes. Phylogenetic analysis of cp genome of M. longirostrata with 11 chloroplast genomes previously reported in the Magnoliaceae shows that M. longirostrata is close to Manglietia megaphylla with high bootstrap value.
Manglietia longirostrata (D. X. Li et R. Z. Zhou ex X. M. Hu, Q. W. Zeng et L. Fu) Sima was combined and stated by Sima Yongkang (Sima et al. Citation2016), Which was described as Magnolia hookeri var. Longirostrata for the long-beaked apex of the follicles and scanning electron micrographs of pollen grains by Hu X. M. & Q. W. Zeng, and they only found one individual in Maocaoping, Malipo County, Yunnan Province, China (Hu et al., 2012). After a careful field study, Sima Yongkang et al. found this species is scattered rarely in southeastern of Yunnan province in China, including Malipo, Yuanyang, and Jingpin county, the altitude range is 900 ∼ 1300 m, and the species was combined and stated in the genus Manglietia in the Magnoliaceae (Sima et al. Citation2016). Here, the annotated chloroplast (cp) genome sequence of M. longirostrata has been assembled and submitted to the GenBank with the accession number MT584886 and we performed a phylogenetic analysis which would benefit the genetic and phylogenetic research within this genus species.
The fresh leaves of M. longirostrata were collected from a tree cultivated in Kunming Arboretum, Yunnan Academy of Forestry & Grassland Science, Yunnan Province of China (25°9′5′′ N, 102°44′45′′ E). The sheets of the vouchered specimen (Sima and Lu Citation2012) are stored at the herbaria of YAF and YCP.
Total genomic DNA was extracted from the fresh leaves using Rapid Plant Genomic DNA Isolation Kit. The extracted DNA was sequenced using the Illumina Miseq platform (Illumina, San Diego, CA). In total, 85.1 M of 150-bp raw reads were retrieved. In order to ensure the quality of information analysis, the original reads must be filtered to get lean reads using Trimmomatic (Bolger et al. Citation2014). Sequencing data were assembled with SPAdes and GapFille (Boetzer and Pirovano Citation2012) was used to supplement the GAP of the contig obtained by stitching. The genome was automatically annotated using Prokka (Seemann Citation2014). OGDRAW v1.3.1 (Greiner et al. Citation2019) was used to generate a physical map of the cp genome.
The length of the complete cp genome sequence of M. longirostrata is 160,049 bp with four sub-regions: 88,098 bp of large single-copy (LSC) region and 18,864 bp of small single-copy (SSC) region are separated by two inverted repeats (IR) regions, each 26,571 bp, akin to other taxa in the family of Magnoliaceae (Liang et al. Citation2020). The overall CG content of the M. longirostrata cp genome is 39.30%. The cp genome contained 123 genes, including 73 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. 15 genes (trnA-UGC, trnE-UUC, trnK-UUU, trnS-CGA, trnL-UAA, trnC-ACA, trnE-UUC, rps16, rps12, rpl2, atpF, ndhB, ndhA, ycf1, and ycf3) contain intron. The annotated genomic sequence was submitted to GenBank under the Accession Number of MT584886.
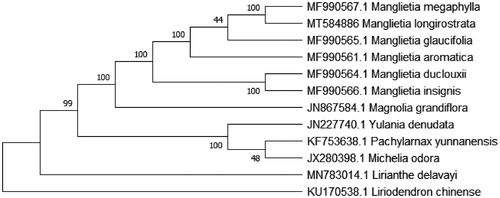
The complete cp genome of 9 reported Magnoliaceae species and one Liriodendron species as an outgroup were downloaded from NCBI GenBank and Lirianthe delavayi’s cp genome sequence from our work before (Liang et al. Citation2020) for the phylogenetic analysis. The combined datasets of 12 species were aligned by Kalign (Madeira et al. Citation2019). A maximum-likelihood (ML) tree was constructed in MEGA X with 1000 bootstrap replications (Kumar et al. Citation2018). The phylogenetic tree reveals that M. longirostrata is most closely related to M. megaphylla with strong bootstrap support () and the genera phylogenetic relationship is almost accord with the phylogenetic tree of Lirianthe coco (Sima et al. Citation2020) and all genera mentioned in this study are monophyletic under the taxonomical system of Magnoliaceae by Sima and Lu (Citation2012). We think that the cp genome resource of M. longirostrata will be valuable for future studies in conservation genetics, taxonomy, phylogeny, and breeding in the Manglietia species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The cpDNA sequence of Manglietia longirostrata of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/genbank/, reference number: MT584886.
Scientific name of the organism in the paper: Manglietia longirostrata Sima.
Geographic location of the specimen: Kunming Arboretum, Yunnan Academy of Forestry & Grassland Science, Yunnan Province of China (25°9′5′′ N,102°44′45′′ E).
Additional information
Funding
References
- Boetzer M, Pirovano W. 2012. Toward almost closed genomes with GapFiller. Genome Biol. 13(6):R56.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes . Nucleic Acids Res. 47(W1):W59–W64.
- Hu XM, Zeng QW, Fu L. 2012. Magnolia hookeri var. longirostrata (Magnoliaceae), a new taxon from Yunnan, China. Ann Bot Fennici. 49(5–6):417–421.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Liang A, Luo W, Li Z, Sima Y, Xu T. 2020. The complete chloroplast genome sequence of Magnoliadelavayi (Magnoliaceae), a rare ornamental and medical tree endemic to China. Mitochondrial DNA Part B. 5(1):883–884.
- Madeira F, Park Y-m, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, et al. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47(W1):W636–W641.
- Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 30(14):2068–2069.
- Sima Y-K, Chen W-H, Shui Y-M. 2016. Diversity and geographical distribution of the Magnoliaceae of Xilong Mountain in South Yunnan, China. In: Shui Y-M, editor Seed plants of Xilong Mountain, the highest mountain in South Yunnan, China. Beijing (China): Science Press; p. 108–130.
- Sima Y-K, Lu SG. 2012. A new system for the family Magnoliaceae. In: Xia NH, Zeng QW, Xu FX, Wu QG, editors. Proceedings of the second international symposium on the family Magnoliaceae. Wuhan (China): Huazhong University Science & Technology Press; p. 55–71.
- Sima Y-K, Wu T, Fu Y, Hao J, Chen S. 2020. Complete chloroplast genome sequence of Lirianthecoco (Loureiro) N. H. Xia & C. Y. Wu (Magnoliaceae), a popular ornamental species, Mitochondrial DNA Part B. Mitochondrial DNA Part B. 5(3):2410–2412.