Abstract
The complete mitochondrial genome (mitogenome) of Polygonia c-aureum, from Yizhou of China, is a circular molecule of 15,202 bp in length, containing 13 protein-coding genes, 2 ribosomal RNAs, 22 transfer RNAs, and a control region. The A + T content of the overall base composition is 80.6%. All of the 13 PCGs are begun with ATN (one with ATC, four with ATT and seven with ATG) as start codon except cox1 using CGA. Ten genes are terminated with TAA as stop codon, cox2, nad4 and nad5 end with an incomplete stop codon (T). The newly sequenced mitogenome will increase the basic information of Nymphalinae phylogenetic research and can help to better understand the phylogenetic status of P. c-aureum.
The subfamily Nymphalinae consists of the tribes Nymphalini, Melitaeini, Kallimini, Victorinini, Junoniini, and probably the Coeini, but the phylogenetic relationships among Nymphalinae remains puzzling (Shi et al. Citation2015; Su et al. Citation2017). Polygonia c-aureum, an important member of this subfamily, are widely distributed in Korea, Japan, Mongolia, Vietnam, Siberia, and in all regions of China except Tibet (Chou Citation1994). In addition, P. c-aureum exhibits seasonal polyphenism in wing color, wing morphology and reproduction (Hiroyoshi et al. Citation2020). Therefore, mitochondrial genome studies of P. c-aureum will contribute to the study of its own population genetics and phylogeny of the subfamily Nymphalinae.
In this study, we sequenced the complete mitogenome of P. c-aureum (GenBank accession No. MT654530). Adult specimens of P. c-aureum were collected in Yizhou, Guangxi Zhuang Autonomous Region, China. The fresh individuals were preserved in 95% ethanol at temperature −20 °C. Total genomic DNA was extracted from the leg of a single individual using a Wizard® Genomic DNA Purification Kit (Promega, Madison, USA) according to the manufacturer’s instructions. The specimens and DNA of these specimens were deposited in the Museum of Insects of Hechi University (Voucher No. L389). The genomic DNA was sequenced using the Hiseq2500 platform (Illumina, San Diego, CA). Mitochondrial genome was assembled by Geneious 9.0.4 (https://www.geneious.com), and annotated using MITOS Web Server (Bernt et al. Citation2013).
The complete mitogenome is 15,202 bp in length and contains 13 protein-coding genes, two ribosomal RNAs, 22 transfer RNAs genes, and a control region (called A + T rich-region). Its gene arrangement and direction are in accordance with other Nymphalid butterflies (Xu et al. Citation2019). The mitogenome has an A + T content of 80.6% (A 40.1%; T 40.5%; C 11.9%; G 7.5%), which is a typical structure of lepidopteran mitogenome (Yang et al. Citation2019). All of the 13 PCGs were begun with ATN (one with ATC, four with ATT and seven with ATG) as start codon except cox1 using CGA. Ten genes were terminated with TAA as stop codon, cox2, nad4 and nad5 ended with an incomplete stop codon (T). The rrnS (776 bp) and rrnL (1366 bp), were located between the trnL1 and AT-rich region, and separated by the trnV gene. 22 tRNA genes ranged in size from 62 to 71 bp. All tRNAs harbor the typical predicted secondary cloverleaf structures except for the trnS1, as seen in all other determined butterflies (Shi et al. Citation2015).
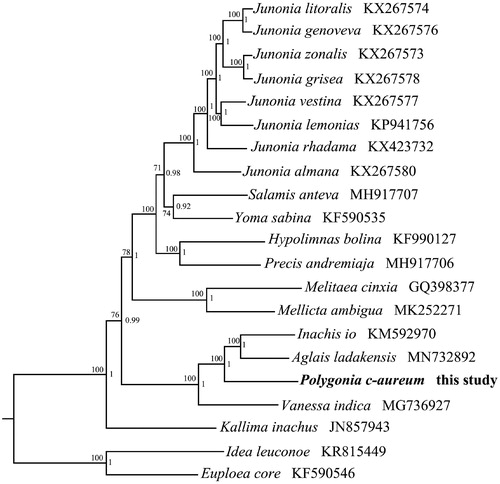
Phylogenetic trees were constructed using the 13 PCGs from 19 Nymphalinae species and two outgroups of Danainae, respectively. Maximum Likelihood (ML) and Bayesian Inference (BI) methods were employed for the dataset to assess whether the datasets were sensitive to the inference methods (Ronquist et al. Citation2012; Stamatakis Citation2014). We used the best-fit partitioning scheme and partition-specific models recommended by PartitionFinder (Lanfear et al. Citation2012). Two phylogenetic analyses using different methods yielded the same topology, and nodal supporting values were always higher for BI tree than for ML tree. The phylogenetic tree indicated that the genus Junnia was a monophyletic group in Nymphalinae. As shown in , P. c-aureum is a sister clade to Aglais ladakensis + Inachis io, then cluster with Vanessa indica. Our analyses indicated that our newly determined mitogenome sequence will be useful for studying population genetics and phylogenetic analysis of this species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in National Center for Biotechnology Information at https://www.ncbi.nlm. nih.gov/nuccore, reference number [MT654530].
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chou I. 1994. Monographia Rhopalocerorum Sinensium. Zhengzhou: Henan Scientific and Technological Press.
- Harvey DJ. 1991. Higher classification of the Nymphalidae, Appen-dix B. In: Nijhout HF, editor. Higher classification of the Nymphalidae, ppendix B. Washington (DC): Smithsonian Institution Press; p. 255–273.
- Hiroyoshi S, Reddy GVP, Mitsunaga T. 2020. Effects of photoperiod and aging on the adult spermatogenesis of Polygonia c-aureum (Lepidoptera: Nymphalidae), in relation to adult diapause. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 206(3):467–475.
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29(6):1695–1701.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Shi QH, Sun XY, Wang YL, Hao JS, Yang Q. 2015. Morphological characters are compatible with mitogenomic data in resolving the phylogeny of nymphalid butterflies (Lepidoptera: Papilionoidea: Nymphalidae). PLOS One. 10(4):e0124349.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Su CY, Shi QH, Sun XY, Ma J, Li CH, Hao JS. 2017. Dated phylogeny and dispersal history of the butterfly subfamily Nymphalinae (Lepidoptera: Nymphalidae). Sci. Rep. 7(8799):1–11.
- Xu C, Pan ZQ, Nie L, Hao JS. 2019. The complete mitochondrial genome of Issoria eugenia (Lepidoptera: Nymphalidae: Heliconiinae). Mitochondr DNA Part B. 4(1):1662–1663.
- Yang M, Song L, Shi YX, Li JH, Zhang Y, Song N. 2019. The first mitochondrial genome of the family Epicopeiidae and higher-level phylogeny of Macroheterocera (Lepidoptera: Ditrysia). Int J Biol Macromol. 136:123–132.