Abstract
Bambusa rigida is a tropical woody bamboo widely distributed in Sichuan and southeastern of China with important economic and ecological values. We performed a complete chloroplast (cp) genome of B. rigida using Illumina NovaSeq PE150 platform. The length of complete cp sequence is 139,500 bp in size with a pair of inverted repeat (IR) regions of 43,587 bp, which separates a large single-copy (LSC) region of 83,036 bp and a small single-copy (SSC) region of 12,875 bp. Plastid genome contains 132 genes, including 84 protein-coding genes, 40 tRNA genes, and 8 rRNA genes. Phylogenetic analysis based on 30 cp genomes indicates that B. rigida is closely related to Bambusa ventricosa in Bambuseae.
Bambusa rigida Keng et Keng f. is a tropical woody bamboo belonging to the family Gramineae (Bambusoideae: Bambuseae) with the rhizome pachymorph without enlonged necks (Li et al. Citation2006). It is widely distributed in Sichuan, Guizhou, Guangxi, Guangdong, Fujian provinces of China with important economic benefits (Liu et al. Citation2013) and ecological functions (P. Zhang et al. Citation2009). Bambusa rigida is a typical medium-sized sympodial bamboo species characterized by high stalk, thick wall, hard material, heavy weight, relatively small diameter, which made it a perfect raw material for paper making as well as for weaving, scaffolding, fence and garden greening (W. Zhang et al. Citation2014; Cao et al. Citation2018). In this study, we reported the complete chloroplast (cp) genome of B. rigida based on Illumina pair-end sequencing data.
Fresh and healthy leaves of B. rigida were collected from Yibin, Sichuan province (28°45′28″N, 104°38′34″E), and dried immediately by silica gel. The voucher specimen was kept at the College of Forestry and Biotechnology, Zhejiang A&F University (Accession No. BR01). Total genomic DNA was extracted using a modified CTAB protocol (Doyle Citation1991). Pair-end library was constructed and sequenced with Illumina NovaSeq PE150 platform (Novogene Bioinformatics Technology Co., Ltd in Beijing, China). The chloroplast genome of B. rigida was assembled by MITObim v1.8 (Hahn et al. Citation2013) and annotated with the PGA (Qu et al. Citation2019).
The complete chloroplast genome of B. rigida (GenBank accession: MT648824) was 139,500 bp in size with a large single-copy (LSC) region of 83,036 bp, a small single-copy (SSC) region of 12,875 bp, and a pair of inverted repeat (IR) regions of 43,587 bp. A total of 132 genes was found in the B. rigida chloroplast genome, including 84 protein-coding genes, 40 tRNA genes, and 8 rRNA genes. The complete genome GC content was 39.42%.
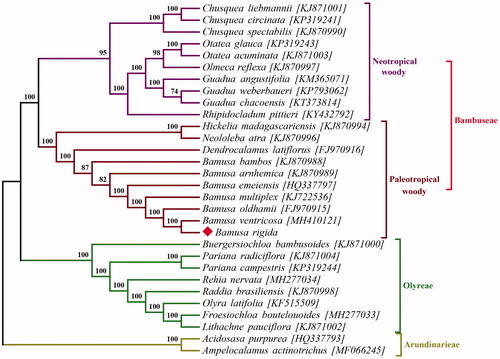
To reveal the phylogenetic relationship of B. rigida within the Gramineae family, a phylogenetic analysis was performed based on 28 complete chloroplast genomes of Bambusoideae with Acidosasa purpurea and Ampelocalamus actinotrichus as outgroups. All of the cp genome data were downloaded from NCBI GenBank database. The sequences were aligned by MAFFT (Katoh and Standley Citation2013). Phylogenetic analysis was conducted by RAxML-NG (Kozlov et al. Citation2019). The bootstrap values were calculated based on 1000 replicates. The phylogenetic tree revealed that B. rigida was most closely related to Bambusa ventricosa with strong support ().
Acknowledgements
The authors thank Hantian Wei’s help in the process of data analysis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study have been deposited in the NCBI database [GenBank accession: MT648824] (https://www.ncbi.nlm.nih.gov/).
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Cao Y, Zeng Y, Bie P, Zhao R, Chen J, Chen X. 2018. A study of the growth of Bambusa rigida in Changning County of Sichuan Province. J Sichuan Forest Sci Technol. 39:120–124.
- Doyle J. 1991. DNA protocols for plants. In: Hewitt GM, editor. Molecular techniques in taxonomy. Berlin: Springer-Verlag; p. 283–293.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21):4453–4455.
- Li DZ, Wang ZP, Zhu ZD, Xia NH, Jia LZ, Guo ZH, Yang GY, Stapleton C. 2006. Bambuseae (Poaceae). In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Beijing and St. Louis: Science Press and Missouri Botanical Garden Press; p. 7–180.
- Liu G, Fan S, Cai C, Zhang D. 2013. Comparative Analysis on the Clonal Growth Characteristics of Bambusa pervariabilis × Dendrocalamopsis daii and B. rigida. Chin Bull Bot. 48:288–294.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50
- Zhang P, Zhang X, Huang L, Liu W, Zhu W, Tang S. 2009. Interception effect of surface runoff on different width Bambusa rigida Keng Riparian Buffer Strips. J Soil Water Conserv. 23:23–27.
- Zhang W, Liu S, Tu S, Jiang B, Wu Z, Hu D, Tu S, Guo X. 2014. Culm form characteristics of Bambusa rigida and the influence of special bamboo fertilizer. J West China Forest Sci. 44:9–14.