Abstract
We sequenced and assembled eight complete plastid genomes from three closely related pleurocarpous moss families: Amblystegium serpens, Campyliadelphus stellatus, Cratoneuron filicinum, Drepanocladus aduncus, and Leptodictyum humile (Amblystegiaceae), Calliergon sarmentosum and Warnstorfia exannulata (Calliergonaceae), and Calliergonella cuspidata (Pylaisiaceae). The newly generated plastid genomes range from 124,256 to 124,819 bp, with two inverted repeat regions (9,624–9,696 bp) separated by a large single-copy region (86,422–86,924 bp) and a small single-copy region (18,430–18,514 bp). All these plastid genomes encode 116 unique genes including 82 protein-coding genes, 30 tRNA genes and four rRNAgenes. The overall GC content is between 28.6%–29.3%. Phylogenetic analysis showed that all Amblystegiaceae species Amblystegium serpens, Campyliadelphus stellatus, Cratoneuron filicinum, Drepanocladus aduncus, Leptodictyum humile, and Sanionia uncinata clustered in one clade, which is sister to the Pylaisiaceae species Calliergonella cuspidata. The two Calliergonaceae species Calliergon sarmentosum and Warnstorfia exannulata form a clade and is sister to Amblystegiaceae and Pylaisiaceae.
The moss family Amblystegiaceae was first recognized in 1885 and was placed close to Hypnaceae (Kindberg Citation1885). Then the family was thoroughly redescribed by Roth (Citation1899). Since the second edition of the book Musci in Natürlichen Pflanzenfamilien (Brotherus Citation1925), Amblystegiaceae was universally accepted as a family. Amblystegiaceae were traditionally circumscribed by their mostly single and long costa in leaf, cylindrical and curved spore capsule, and their preference for moist biotope (Hedenäs and Vanderpoorten Citation2007). Molecular phylogenetic studies based on chloroplast (trnL-trnF and atpB-rbcL) and nuclear (ITS) markers have provided strong evidence that the family should be splitted into Amblystegiaceae and Calliergonaceae (Hedenäs et al. Citation2005; Vanderpoorten et al. Citation2002a, Citation2002b, Citation2003), this treatment has been followed by the classification of the Bryophyta of Goffinet & Buck (Citation2004), Goffinet et al. (Citation2008), and Frey & Stech (Citation2009). Considering the complicated relationship of Amblystegiaceae, we selected eight species () belonging to eight genera of traditional Amblystegiaceae based on morphological classification (Vitt Citation1984), and assembled and annotated their complete plastid genomes.
Table 1. Voucher information of the eight samples.
The total DNA was extracted using the modified CTAB method (Forrest et al. Citation2011). Genome sequencing was performed using the BGISEQ platform (BGI, Shenzhen, China), and about 3 Gb raw sequence data were generated for each sample. The sequence reads were assembled using GetOrganelle (Jin et al. Citation2018), which relies on SPAdes (Bankevich et al. Citation2012), Bowtie2 (Langmead and Salzberg Citation2012), and BLAST+ (Camacho et al. Citation2009). For the annotation, the program PGA (Qu et al. Citation2019) was used, with the plastid genome of Sanionia uncinata (NC_025668.1; Park et al. Citation2018), an Amblystegiaceae specie as the reference.
The newly generated plastid genomes are between 124,256 to 124,819 bp in size, and with two inverted repeat regions (9,624–9,696 bp) separated by a large single-copy region (86,422–86,924 bp) and a small single-copy region (18,430–18,514 bp). The plastid genome each encodes 116 unique genes including 82 protein-coding genes, 30 tRNA genes, and four rRNA genes. Among these, five tRNA genes (trnA-UGC, trnI-GAU, trnN-GUU, trnR-ACG, and trnV-GAC) and four rRNA genes are duplicated in the IR regions. A total of 16 genes harbor intron, 14 genes (atpF, ndhA, ndhB, rpoC1, rps12, rpl16, rpl2, ycf66, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) contain one intron, and two genes (ycf3 and clpP) contain two introns. The overall GC content of these plastid genomes is between 28.6% to 29.3%.
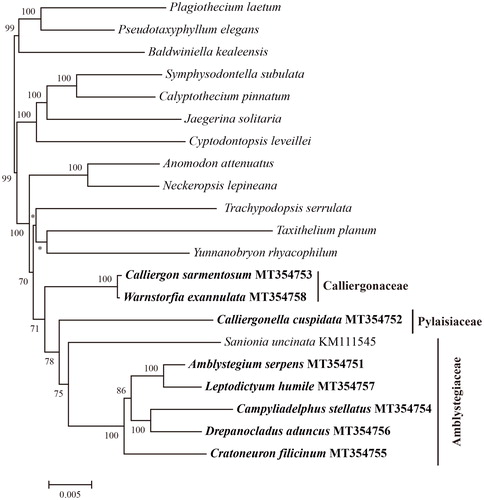
To reconstruct the phylogenetic relationships of these eight species within the Hypnales, we composed a data matrix of plastid genes derived from the newly seuqenced plastid gneomes, the plastid genome of Sanionia uncinata (Park et al. Citation2018), and plastid protein-coding genes from 12 genera of Hypnales (Liu et al. Citation2019). A total of 82 plastid protein-coding genes were extracted, and aligned with MAFFT v7.355 (Nakamura et al. Citation2018). The Maximum Likelihood tree was calculated under the parameter of PROTGAMMAAUTO using RAxML v8.2.11 (Stamatakis Citation2014). The resulted phylogenetic tree reveals the Amblystegiaceae species Amblystegium serpens, Campyliadelphus stellatus, Cratoneuron filicinum, Drepanocladus aduncus, Leptodictyum humile, and Sanionia uncinata cluster together, and is sister to the Pylaisiaceae Calliergonella cuspidata, and the Calliergonaceae species Calliergon sarmentosum and Warnstorfia exannulata form a clade and is sister to Amblystegiaceae and Pylaisiaceae (). This result is consisitent with former studies of Hedenäs et al. (Citation2005), Hedenäs & Vanderpoorten (Citation2007), and Vanderpoorten et al. (Citation2002a, Citation2002b), supporting the traditional family Amblystegiaceae should be splitted into Amblystegiaceae and Calliergonaceae.
Figure 1. The maximum likelihood tree of 21 Hypnales species based on 82 plastid protein-coding genes. The numbers above the branches are bootstrap support values, * indicate bootstrap is < 50. The newly seqeunced eight species Amblystegium serpens, Campyliadelphus stellatus, Cratoneuron filicinum, Drepanocladus aduncus, Leptodictyum humile, Calliergon sarmentosum, Warnstorfia exannulata, and Calliergonella cuspidata are in bold. The data of the species without GenBank accession numbers were retrieved from the study of Liu et al. (Citation2019). The classfication followed Goffinet et al. (Citation2008) and Paulo et al. (Citation2018).
Acknoledgement
The authors thank Dr. Shan-Shan Dong and Bin Tian for asistance in analysis and lab work.
Disclosure statement
The authors declare no conflicts of insterest.
Data availability statement
The annotated plastid genomes have been deposited in the GenBank database (accession number MT354751–MT354758). The raw genomic NGS read data for assembling the plastid genomes have been deposited in the NCBI Sequence Read Archive (SRA; accession no. SRP260466; https://www.ncbi.nlm.nih.gov/sra/?term=SRP260466).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Brotherus VF. 1925. Musci. In: Engler A, editor. Die Natürlichen Pflanzenfamilien, Vol. 11.Leipzig: Verlag von Wilhelm Engelmann; p. 1–542.
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics. 10:421.
- Forrest LL, Wickett NJ, Cox CJ, Goffinet B. 2011. Deep sequencing of Ptilidium (Ptilidiaceae) suggests evolutionary stasis in liverwort plastid genome structure. Plecevo. 144(1):29–43.
- Frey W, Stech M. 2009. Bryophyta (Musci, Mosses). In: Frey W, editor. Syllabus of plant families A. Engler’s syllabus der pflanzenfamilien. Part 3. Bryophytes and seedless vascular plants. 13th ed. Stuttgart, Germany: Gebr. Borntraeger Verlagsbuchhandlung; p. 220–221.
- Goffinet B, Buck WR, Shaw AJ. 2008. Morphology, anatomy, and classification of the bryophyta. In: Goffinet B and Shaw AJ, editors. Bryophyte biology. 2nd ed. Cambridge University Press, Cambridge; p. 53–138.
- Goffinet B, Buck WR. 2004. Systematics of the Bryophyta (mosses): from molecules to a revised classification. In Goffinet B, Hollowell V, and Magill R, editor. Molecular systematics of bryophytes. Missouri Botanical Garden Press; p. 205–239.
- Hedenäs L, Oliván G, Eldenäs P. 2005. Phylogeny of the Calliergonaceae (Bryophyta) based on molecular and morphological data. Plant Syst Evol. 252(1–2):49–61.
- Hedenäs L, Vanderpoorten A. 2007. The Amblystegiaceae and Calliergonaceae. In: Newton AE and Tangney RS, editor. Pleurocarpous mosses: systematics and evolution. London: Taylor & Francis; p. 163–176.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. bioRxiv 256479..
- Kindberg NC. 1885. Table analytique des mousses pleurocarpes européennes. Revue Bryologique. 12:24–31.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359.
- Liu Y, Johnson MG, Cox CJ, Medina R, Devos N, Vanderpoorten A, Hedenäs L, Bell NE, Shevock JR, Aguero B, et al. 2019. Resolution of the ordinal phylogeny of mosses using targeted exons from organellar and nuclear genomes. Nat Commun. 10(1):1485.
- Nakamura T, Yamada DK, Tomii K, Katoh K. 2018. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics. 34(14):2490–2492.
- Park M, Park H, Lee H, Lee BH, Lee J. 2018. The complete plastome sequence of an antarctic bryophyte Sanionia uncinata (Hedw.) loeske. IJMS. 19(3):709.
- Paulo EC, Micheline CS, Diego KH, Juan G, María TG, Diana RP, Michael S. 2018. Pylaisiaceae Schimp. (Bryophyta) revisited. J Bryol. 40(3):1–14.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Roth G. 1899. Uebersicht über die familie der Hypnaceen. Hedwigia, Beiblatt. 1:3–8.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Vanderpoorten A, Goffinet B, HedenäS L, Cox CJ, Shaw AJ. 2003. A taxonomic reassessment of the Vittiaceae (Hypnales, Bryopsida): evidence from phylogenetic analyses of combined chloroplast and nuclear sequence data. Plant Syst Evol. 241(1–2):1–12.
- Vanderpoorten A, Hedenäs L, Cox C, Shaw AJ. 2002a. Circumscription, classification, and taxonomy of Amblystegiaceae (Bryopsida) inferred from nuclear and chloroplast DNA sequence data and morphology. Taxon. 51(1):115–122.
- Vanderpoorten A, Hedenäs L, Cox C, Shaw AJ. 2002b. Phylogeny and morphological evolution of the Amblystegiaceae (Bryopsida). Mol Phylogenet Evol. 23(1):1–21.
- Vitt DH. 1984. Classification of the Bryopsida. In: Schuster RM, editor. New manual of bryology, Vol. 2. Miyazaki: Hattori Botanical Laboratory; p. 696–759.
