Abstract
New mitochondrial genomes of Pseudogobio guilinensis, P. giganteus, and P. anderssoni have the length of 16,605, 16,606, and 16,609 bp with A + T bias. Inferred phylogeny shows that P. guilinensis occupies basal position. P. esocinus and P. anderssoni+P. longirostris are sister groups, and they together are a sister taxa of P. giganteus.
Pseudogobio fishes (Cypriniformes: Gobionidae) are a group of freshwater small-sized fishes (<15 cm), and they are distributed in East Asia (Tominaga and Kawase Citation2019; Fu Citation2020). The genus Pseudogobio has been revised in a recent study (Fu Citation2020), and they are comprised of eight valid nominated species and two cryptic species. Mitochondrial genomes of three species P. guilinensis, P. giganteus, and P. anderssoni have been determined in this study, and they would be used to delimit Pseudogobio species and to infer their phylogeny.
Pseudogobio guilinensis (voucher FDZM-PGuPingL20170716-01) is collected from the Pingle County, China (24.64°N, 110.65°E), P. anderssoni (FDZM-PAAnY20180728-01) from the Anyi County, China (28.84°N, 115.56°E), and P. giganteus (FDZM-PGiXYing20170921-01) from the Haikou City, China (24.64°N, 110.65°E). These specimens have been deposited in the Zoological Museum of Fudan University (FDZM). The Sanger sequencing is used to obtain mitochondrial genomes.
Three new mitochondrial genomes consist of 13 protein-coding genes, two rRNA genes, 22 tRNA genes, and one control region. Their lengths are 16,605, 16,606, and 16,609 bp with A + T bias of base compositions 56.8, 57.8, and 57.5%. Among 13 protein-coding genes, GTG is the start codon of COI gene, and ATG is start codons of the remaining 12 genes. Four kinds of stop codons are revealed to include TAG (ATP8, ND5), TAA (ND1, COX1, ATP6, ND4L, ND6), T— (COX2, COX3, CYTB), and TA– (ND2, ND3, ND4). Six pairs of adjoining genes show 1–7 bp of gene overlaps and 13 pairs of adjoining genes show 1–31 bp of gene intervals. Patterns in gene arrangements, codon uses, gene overlaps, and gene intervals are consistent with published fish mitochondrial genomes (Li et al. Citation2018; Tong and Fu Citation2019; Zhang and Fu Citation2019).
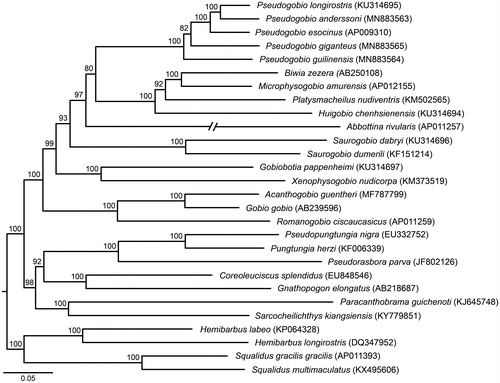
Phylogenetic relationships between Pseudogobio fishes and their close relatives are inferred using five partitions (each codon of all protein-coding genes, 12S + 16S rRNA genes, and all tRNA genes) of 28 mitochondrial genomes in the software IQ-TREE 1.6.2 (Nguyen et al. Citation2015). Our tree () reveals that the genus Pseudogobio is a monophyletic group. P. guilinensis is located in basal position. P. esocinus and P. anderssoni + P. longirostris are sister groups, and they together are a sister taxa of P. giganteus.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
Three new mitochondrial genomes have been released in the GenBank with accession numbers as follows: MN883563 (https://www.ncbi.nlm.nih.gov/nuccore/MN883563); MN883564 (https://www.ncbi.nlm.nih.gov/nuccore/MN883564); MN883565 (https://www.ncbi.nlm.nih.gov/nuccore/MN883565).
Additional information
Funding
References
- Fu JW. 2020. Studies on phylogeny and speciation of the genus Pseudogobio and genealogical divergence and colonization history of Pseudogobio anderssoni [M.S. thesis]. Shanghai, China: Fudan University.
- Li YH, Cao K, Fu CZ. 2018. Ten fish mitogenomes of the tribe Gobionini (Cypriniformes: Cyprinidae: Gobioninae). Mitochondrial DNA B. 3(2):803–804.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Tominaga K, Kawase S. 2019. Two new species of Pseudogobio pike gudgeon (Cypriniformes: Cyprinidae: Gobioninae) from Japan, and redescription of P. esocinus (Temminck and Schlegel 1846). Ichthyol Res. 66(4):488–508.
- Tong J, Fu CZ. 2019. Four complete mitochondrial genomes of Saurogobio fishes (Cypriniformes: Gobionidae). Mitochondrial DNA B. 4(2):2175–2176.
- Zhang X, Fu CZ. 2019. Two complete mitochondrial genomes of Paraleucogobio fishes (Cypriniformes: Gobionidae). Mitochondrial DNA B. 4(2):2173–2174.