Abstract
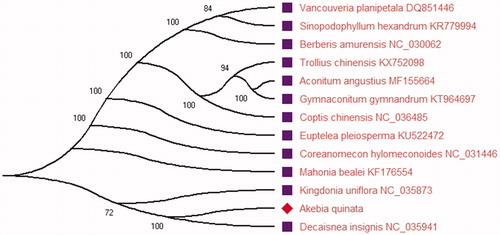
Akebia quinata is an herb medicine plant in traditional oriental medicine in China. In this current study, we had assembled the complete chloroplast genome of A. quinata. The complete chloroplast genome sequence of A. quinata has a total 157,817 bp in size and has a typical quadripartite structure, which is the same as other plant species chloroplast genome. It contained a large single-copy (LSC) region of 86,544 bp, a small single-copy (SSC) region of 18,989 bp and two inverted repeat (IR) regions of 26,142 bp. The complete chloroplast genome sequence contains 132 genes, including 87 encoding genes, 37 transfer RNA genes, and eight ribosomal RNA genes. One of IR region harbors seven protein encoding genes, seven tRNA genes, and four rRNA genes. Phylogenetic analysis result showed that A. quinata is closely related to Decaisnea insignis of the family Lardizabalaceae using the maximum-likelihood (ML) method in this study.
Akebia quinata is an herb medicine plant in traditional oriental medicine in China. Akebia quinata (Lardizabalaceae) is a creeping woody vine that is widely distributed in East Asia, including China, Korea, and Japan (Sun et al. Citation2018). Akebia quinata is the cultivation of woods, hedges, and thickets in mountainous areas, but also is at forest margins along streams, scrub on mountain slopes at elevations of 300–1500 m in China. Akebia quinata has long time been widely used as an antiphlogistic, a diuretic, and an analgesic in many East Asia countries (Kitaoka et al. Citation2009). Akebia quinata is one of the important herbal medicines in China, but fewer genomic information and study has been reported of this species. Over here, we had assembled the complete chloroplast genome of A. quinata with the good knowledge of chloroplast genome information of this species that can provide the traditional Chinese medicine study and utilization in the future.
The chloroplast DNA of A quinata was extracted from its fresh stems which were sampled in the market of herb near the Third Hospital of Nanchang (28.66 N, 115.90E). The corresponding voucher herbarium specimen was stored at the Third Hospital of Nanchang and the accession number was THNC-03. We controlled and removed the sequences using FastQC (Andrews Citation2015) after which the chloroplast genome DNA was purified and sequenced. The chloroplast genome was constructed with de novo assembly and mapped using NOVOPlasty (Dierckxsens et al. Citation2017). The chloroplast genome of A. quinata gene annotation was done using Geneious (Kearse et al. Citation2012). All the genes were analyzed using CPGAVAS (Liu et al. Citation2012) and combined with the online alignment tool Blastx and ORF Finder from the NCBI web (http://www.ncbi.nlm.nih.gov/). The tRNA genes were predicted using the online sites tRNA-scan (Lowe and Chan Citation2016). The circular chloroplast genome map was finished using OrganellarGenomeDRAW (Greiner et al. Citation2019) (http://ogdraw.mpimp-golm.mpg.de/).
The chloroplast genome sequence of A. quinata had been submitted to NCBI and the accession number was NK9912251. The complete chloroplast genome sequence of A. quinata has a total 157,817 bp in size and has a typical quadripartite structure, which is the same as other plant species chloroplast genome. It contained a large single-copy (LSC) region of 86,544 bp, a small single-copy (SSC) region of 18,989 bp and two inverted repeat (IR) regions of 26,142 bp. The overall nucleotide composition of chloroplast genome sequence is A 30.3% (47,841 bp), T 31.0% (48,952 bp), C 19.7% (31,052 bp), G 19.0% (29,972 bp), and the total G + C content of 38.7%. The complete chloroplast genome sequence contains 132 genes, including 87 encoding genes, 37 transfer RNA genes, and eight ribosomal RNA genes. One of IR region harbors seven protein encoding genes (rpl2, rpl23, ycf2, ndhB, rps7, rps12, and ycf1), seven tRNA genes (trnI-CAU, trnL-CAA, trnV-GAC, trnIGAU, trnA-UGC, trnR-ACG, and trnN-GUU), and four rRNA genes (rrn16, rrn23, rrn4.5, and rrn5).
The phylogenetic analyses were carried out using the complete chloroplast genome sequences of A. quinata and 12 plant species reported the complete chloroplast genome. Before constructing the phylogenetic tree, sequence alignment of all the species was performed with MEGA X (Kumar et al. Citation2018). The maximum-likelihood (ML) phylogenetic tree was constructed using MEGA X. The ML phylogenetic tree was inferred with strong support and used the bootstrap values from 2000 replicates at all the nodes. The ML tree was drawn and edited using MEGA X. Phylogenetic analysis result () showed that A. quinata is closely related to Decaisnea insignis of the family Lardizabalaceae using the ML method in this study. The good knowledge of chloroplast genome information of this species can provide the traditional Chinese medicine study and utilization in the future.
Data openly available in a public repository that issues datasets with DOIs
The data that support the findings of this study are openly available in Akebia quinata at http://doi.org/10.1080/23802359.2020.1797557, reference number [NK9912251].
Disclosure statement
The authors have declared that no competing interests exist.
Data availability statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
References
- Andrews S. 2015. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kitaoka F, Kakiuchi N, Long CF, Itoga M, Mitsue A, Mouri C, Mikage M. 2009. Molecular characterization of Akebia plants and the derived traditional herbal medicine. Biol Pharm Bull. 32(4):665–670.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Sun HP, Seol J, Lee SW, Sun DP, Sung YY, Kim HK. 2018. Akebia quinata decaisne aqueous extract acts as a novel anti fatigue agent in mice exposed to chronic restraint stress. J Ethnopharmacol. 222:270–279.