Abstract
The nearly complete mitochondrial genome of Eutomostethus vegetus Konow, 1898 was high-quality assembled. Several gene rearrangement events were observed in IQM gene cluster by comparing with the inferred insect ancestral mitochondrial genome. Phylogenetic analysis demonstrated the position of Eutomostethus vegetus in the Tenthredinidae and showed that Blennocampinae is a sister group to Fenusinae.
Eutomostethus Enslin, 1914 is a member of Tomostethini of Blennocampinae (Wei and Nie Citation1998). The genus is the largest one of Blennocampinae, and about 105 valid species have been described in the world. Species of the genus are the common pest of the plants of Gramineae, including Poa, Juncus, and Phyllostachys, etc. Here, we described the nearly complete mitochondrial genome of E. vegetus Konow, 1898, as the first one of Blennocampinae to advance our understanding of its phylogenetic status within Tenthredinidae.
Specimen (CSCS-Hym-MC0184) was deposited in the Asia Sawfly Museum, Nanchang (ASMN). It was collected at Mt. Lu (29.57°N, 115.96°E), Jiangxi province in China, in April 2019. Whole genomic DNA was extracted from the thorax muscle of specimen (SAMN15311680) using the DNeasyR Blood & Tissue Kits (Qiagen, Valencia, CA). Genomic DNA was sequenced by the high-throughput Illumina Hiseq 4000 platform, yielding a total of 48,582,996 raw reads (SRR12057813). One contig was generated by MitoZ (Meng et al. Citation2019), and then was thoroughly checked by assembly using Hemibeleses tianmunicus (unpublished) and Periclista xanthosomata (unpublished) as references (coverage were 2,787,422 and 3954, respectively). We used part of nad2 (100 bp), trnI (66 bp), trnQ (70 bp) as reference sequences to examine the longest interval region further. By consistently obtaining similar coverage of the assembly contigs, we were able to confirm the 680 bp non-coding region between trnQ and trnI. Annotations were generated in MITOS web server (Bernt et al. Citation2013) and revised when necessary. The accession number of BioProject is PRJNA640463.
The nearly complete mitochondrial genome of E. vegetus (MT663219) was 16,345 bp in length and biased toward A and T, with an 80.8% A + T content. The mitogenome of E. vegetus contains 13 protein-coding genes (PCGs), 22 tRNA genes, and 2 rRNA genes. Seven tRNAs (trnC, trnY, trnF, trnH, trnP, trnV, trnL1), two rRNAs, and four PCGs (nad1, nad4, nad4L and nad5), were located on the N-strand, while the J-strand encoded the remaining. Compared with the ancestral gene arrangement of insects (Boore Citation1999), trnI translocated to upstream of nad2, and trnM and trnQ swapped positions. Most PCGs initiated by ATN codons and all PCGs ended with TAA stop codons. There were five gene overlaps among trnW-trnC (8 bp), atp8-atp6 (7 bp), atp6-cox3 (1 bp), nad6-cob (1 bp), trnS-nad1 (2 bp). A total of 1251 bp of intergenic spacer sequences were found in 20 locations and varied in size from 1 to 680 bp, with the longest located between trnQ and trnI.
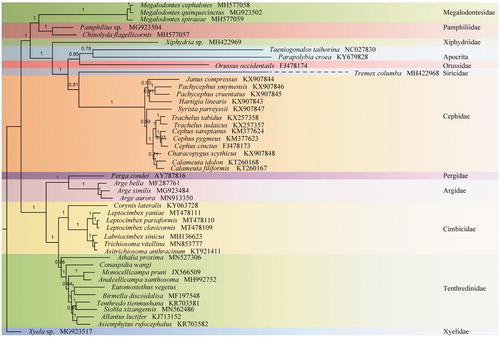
Ten unsaturated amino acid sequences (nad3, atp8, and nad4L were excluded) of 45 Hymenoptera were aligned by TranslatorX (Abascal et al. Citation2010) and concatenated with SequenceMatrix v1.7.8 (Vaidya et al. Citation2011). The phylogenetic trees were constructed using Bayesian inference (BI) with PhyloBayes (Lartillot et al. Citation2009) under the MtArtCAT model in CIPRES (Miller et al. Citation2010) (). It revealed Blennocampinae represented by E. vegetus is a sister group of Fenusinae which was represented by Birmella discoidalisa (MF197548) in Tenthredinidae. All related files have been uploaded to figshare (https://figshare.com/account/home#/projects/83588).
References
- Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38(Web Server issue):W7–W13.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Lartillot N, Lepage T, Blanquart S. 2009. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 25(17):2286–2288.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE), 12 Nov. 2010, New Orleans, LA pp 1–8.
- Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 27(2):171–180.
- Wei M, Nie H. 1998. Generic list of Tenthredinoidea in new systematic arrangement with synonyms and distribution data. J Central South Forestry Univ. 18(3):23–31.