Abstract
In the present study, we sequenced and analyzed the complete mitochondrial genome of the hydrozoan jellyfish Spirocodon saltatrix. The mitochondrial genome was a linear form (15,752 bp long, 70.4% AT), consisting of 13 protein coding genes (cox1, cox2, atp8, atp6, cox3, nad2, nad5, nad6, nad3, nad4L, nad1, nad4, and cytB), two tRNAs (tRNA-Met and tRNA-Trp), and two rRNAs (12S and 16S). Mitochondrial gene arrangement of the S. saltatrix was completely identical to already-known mitochondrial genomes of hydrozoans. Molecular phylogenetic analysis using 13 protein-coding genes showed that S. saltatrix was closely related to the hydrozoan Clava multicornis.
Hydrozoa is one of the five representative classes in the subphylum Medusozoa, and it consists of more than 90% species of medusozoans (Daly et al. Citation2007). In addition, hydrozoans show the most morphological diversity in the Medusozoa (Kayal et al. Citation2015). Recent molecular studies have improved our understanding of their phylogeny (Kayal et al. Citation2015), but many relationships among the sibling species in hydrozoans remain largely unknown.
The hydrozoan jellyfish Spirocodon saltatrix Tilesius 1818 (Cnidaria; Hydrozoa; Anthoathecata) is a single species within the genus (Schuchert Citation2020). Until now, there is no molecular data for this species, excluding 5S ribosomal RNA sequence (GenBank No. K02349). Here, we determined the complete mitochondrial genome of S. saltatrix, and investigated its genomic structures and molecular phylogenetic relationships to other hydrozoans using their genomes.
Environmental specimens of S. saltatrix were collected from Masan Bay (35°09′06.3″N, 128°35″54.3″E) in South Korea, on 27 March 2013. Total genomic DNA (gDNA) was extracted from umbrella tissue using the cetyltrimethylammonium bromide (CTAB) DNA extraction method (Ausubel et al. Citation1989) and the remaining part was stored in the Department of Biotechnology, Sangmyung University, Korea (Accession No. AZ 9-3). Next generation sequencing (NGS) was subjected to the gDNA (Miseq, Illumina, USA), and paired end reads of mitochondrial genome sequences were assembled and annotated using MITObim (Hahn et al. Citation2013), MITOS (Bernt et al. Citation2013), and Geneious 9.1.3 (Geneious, Auckland, New Zealand), respectively. A molecular phylogeny tree was reconstructed using concatenated amino acid sequences of 13 protein-coding genes (PCGs) in MEGA X (Kumar et al. Citation2018). The method of the phylogenetic tree has been described in our previous study (Karagozlu et al. Citation2019).
The complete mitochondrial genome of S. saltatrix sequence was 15,752 bp in length (GenBank No. MT150265; 30.4% A, 40% T, 14% G, and 15.6% C). The genome contained 13 PCGs (cox1, cox2, atp8, atp6, cox3, nad2, nad5, nad6, nad3, nad4L, nad1, nad4, and cytB), two tRNAs (tRNA-Met and tRNA-Trp), and two rRNAs (12S, 16S rRNA). In addition, it had a long non-coding region (656 bp; 17.4% A, 17.8% T, 28.4% C, and 36.4% G) between cox1 and 16S rRNA. The 17 mitochondrial genes arrangement of S. saltatrix was completely identical to those of Anthoathecata hydrozoans, including Clava multicornis (JN700935), Hydra sinensis (JX089978), Hydra oligactis (EU237491), and Turritopsis dohrnii (KT020766). The arrangement, however, was different to other Trachylinae hydrozoans, in which 16S rRNA encoded only in minority strand. Mitochondrial PCGs of S. saltatrix used a single start codon (ATG) and three stop codons (TAA, TAG, and TGA). The most frequent codon in the genome was TTT (409 times, 7.8%).
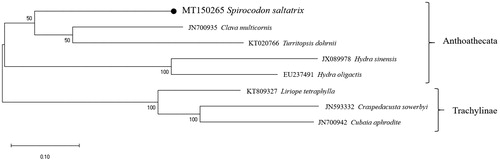
Molecular phylogeny tree () using proteins of 13 mitochondrial PCGs of eight hydrozoan species showed that all Anthoathecata species were clustered together, and the S. saltatrix was the closet to Clava multicornis, followed by T. dohrnii. The present study provides additional mitochondrial genome data of the hydrozoans for understanding the molecular genetic basis of hydrozoans.
Figure 1. Molecular phylogeny of hydrozoans. The tree was reconstructed using concatenated amino acid sequences of 13 mitochondrial protein-coding genes and the maximum-likelihood (ML) algorithm with the JTT matrix-based model in MEGA X software. Bootstrap proportions (BP) of the 1000 times replications were incorporated into the ML tree. The branch lengths are proportional to the scale given. Spirocodon saltatrix determined here represents with a black dot.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The data that support the findings of this study are openly available in GenBank with the accession number MT150265 (https://www.ncbi.nlm.nih.gov/nuccore/ MT150265).
Additional information
Funding
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1989. Current protocols in molecular biology. New York: John Wiley and Sons; p. 4648.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Daly M, Brugler MR, Cartwright P, Collins AG, Dawson MN, Fautin DG, France SC, Mcfadden CS, Opresko DM, Rodriguez E, Romano SL, Stake JL. 2007. The phylum Cnidaria: A review of phylogenetic patterns and diversity 300 years after Linnaeus. Zootaxa 1668:127-182.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:129.
- Karagozlu MZ, Seo Y, Ki JS, Kim CB. 2019. The complete mitogenome of brownbranded moon jellyfish Aurelia limbata (Cnidaria, Semaeostomeae, Ulmaridae) with phylogenetic analysis. Mitochondrial DNA B. 4(1):1875–1876.
- Kayal E, Bentlage B, Cartwright P, Yanagihara AA, Lindsay DJ, Hopcroft RR, Collins AG. 2015. Phylogenetic analysis of higher-level relationships within Hydroidolina (Cnidaria: Hydrozoa) using mitochondrial genome data and insight into their mitochondrial transcription. PeerJ. 3:e1403.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Schuchert P. 2020. World Hydrozoa Database. Spirocodon saltatrix (Tilesius, 1818). World Register of Marine Species. [accessed 2020 May15]. Available at: https://www.marinespecies.org/aphia.php?p=taxdetails&id=291064. on
