Abstract
We generated the complete chloroplast genome sequence of Sinomenium acutum, a species of the Menispermaceae family, and characterized from the de novo assembly of Illumina HiSeq paired-end sequencing data. The total length of the chloroplast genome of S. acutum was 162,787 bp with a large single-copy (LSC) region of 91,430 bp, a small single-copy (SSC) region of 21,245 bp, and a pair of identical inverted repeat regions (IRs) of 25,056 bp. The total of 131 genes were annotated in the chloroplast genome of Sinomenium acutum, including 85 protein-coding genes, 38 transfer RNA (tRNA) genes, and 8 ribosomal RNA (rRNA) genes. The phylogenetic analysis of S. acutum with 18 related species revealed the closest taxonomical relationship with Menispermum dauricum in the Menispermaceae family.
Sinomenium acutum, also known as Chinese moonseed, is a species belonging to the Menispermaceae family which is native to east Asia (Ortiz et al. Citation2007). The root of Sinomenium acutum has been used as medicinal material in China and Korea due to its treatment effect on anti-inflammatory (Zhao et al. Citation2015; Kim et al. Citation2018) and rheumatic disease (Lyu et al. Citation2018). Still many medicinal plants, including Sinomenium acutum, are lack of DNA barcode markers and we generated and the complete chloroplast genome sequence of S. acutum, using next generation sequencing for future resource for marker development. In addition, the sequence of chloroplast genome of Sinomenium acutum would assist future study on genetic diversity and conservation of Menispermaceae speceis.
The plant sample of Sinomenium acutum was maintained and collected from Medicinal Herb Garden, College of Pharmacy, Seoul National University (http://snuherb.snu.ac.kr/) in Goyang, Korea (37°42′44.9″N, 126°49′08.0″E). Total genomic DNA from leaves was used to construct the genomic library for Illumina paired-end (PE) sequencing and also deposited in National Institute of Biological Resources (42 Hwangyeong-ro, Seo-gu, Incheon, 22689, Korea) with collection number of NIBRGR0000622205. The high quality HiSeq reads (>Q30) were assembled by CLC Genomics Workbench (ver. 10.0.1, CLC QIAGEN), followed by manual curation through PE reads mapping (Kim et al. Citation2015). Annotation of the complete chloroplast genome was performed with GeSeq and manual corrections (Tillich et al. Citation2017). The complete chloroplast genome sequence of S. acutum was submitted to GenBank with the accession number of MN626719.
Total length of complete chloroplast genome of S. acutum was 162,787 bp with 37.8% of G + C content, comprising a large single copy (LSC) region of 91,430 bp, a small single copy (SSC) region of 21,245 bp, and a pair of inverted repeat (IRa and IRb) regions of 25,056 bp. The genome contained 131 genes including 85 protein-coding genes, 38 tRNA genes, and 8 rRNA genes.
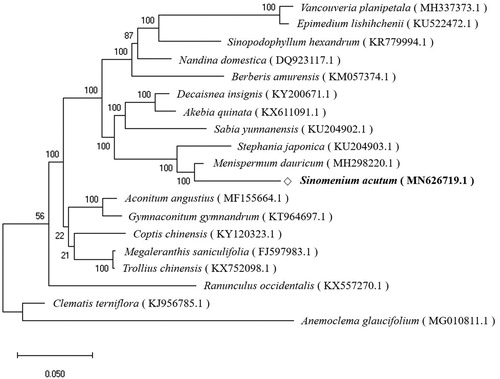
To validate the phylogenetic relationship, the complete chloroplast genome sequences of S. acutum and those from 18 related species were aligned using MAFFT (ver. 7.271) (Katoh et al. Citation2002), followed by phylogenetic tree construction obtained from a Maximum Likelihood (ML) analysis with 1,000 bootstraps using MEGA 7.0 (Kumar et al. Citation2016). The phylogenetic tree exhibited the close relationship of S. acutum with Menispermum dauricum in the family of Menispermaceae ().
Acknowledgements
We appreciate Mr. Sangil Han and Medicinal Herb Garden, College of Pharmacy, Seoul National University, Korea, for providing us plant material of Sinomenium acutum.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number of MN626719.
Additional information
Funding
References
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066.
- Kim K, Lee S-C, Lee J, Yu Y, Yang K, Choi B-S, Koh H-J, Waminal NE, Choi H-I, Kim N-H, et al. 2015. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655.
- Kim TW, Han JM, Han YK, Chung H. 2018. Anti-inflammatory effects of Sinomenium acutum extract on endotoxin-induced uveitis in Lewis rats. Int J Med Sci. 15(8):758–764.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lyu HN, Zeng KW, Cao NK, Zhao B, Jiang Y, Zhao PF. 2018. Alkaloids from the stems and rhizomes of Sinomenium acutum from the Qinling Mountains, China. Phytochemistry. 156:241–249.
- Ortiz RD, Kellogg EA, Van Der Werff H. 2007. Molecular phylogeny of the moonseed family (Menispermaceae): implications for morphological diversification. Am J Bot. 94(8):1425–1438.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Zhao Z, Xiao J, Wang J, Dong W, Peng Z, An D. 2015. Anti-inflammatory effects of novel sinomenine derivatives. Int Immunopharmacol. 29(2):354–360.