Abstract
Acer nikoense (Sapindaceae: Acer) is a deciduous tree, belonging to the Ser. Grisea of Sect. Trifoliata. Its complete genome sequence was obtained using genome Illumina pair-end sequencing data. It had a typical quadripartite structure with 155,952 bp in length, consisting of a large single-copy region (85,720 bp) and a small single-copy region (18,072 bp), as well as a pair of inverted repeats (26,080 bp). The total GC content was 37.9%. A total of 113 unique genes were annotated, including 30 tRNAs, 4 rRNAs, and 79 protein-coding genes. The phylogenetic analysis indicated that A. nikoense and A. triflorum were the most closely related.
Acer nikoense is an attractive deciduous arbor, which belongs to the Ser. Grisea of Sect. Trifoliata in the genus Acer of Sapindaceae. Being one of the ornamental maples, it is naturally distributed in northern Jiangxi Province, southern Anhui Province, western Hubei Province and Japan (Wu et al. Citation2008). According to the International Union for Conservation of Nature (IUCN), it was near threatened (NT) (Wang and Xie Citation2004). It is a lesser-known species, and the studies were mainly focused on propagation, chemical composition, and medicinal value (Kurimoto et al. Citation2016; Kim et al. Citation2017). Till now, the chloroplast genomes of A. griseum (Wang et al. Citation2017) and A. triflorum (Xia et al. Citation2020) which also belong to the Ser. Grisea, were defined, but the information of the complete chloroplast genome of A. nikoense is still lacking. Here, the complete cp genome of A. nikoense was sequenced and assembled to clarify its taxonomic status and lay a foundation for further study on population genetics.
Healthy fresh leaves of A. nikoense were collected from a wild individual maple from the Ningbo City, Zhejiang Province, China (N29°49′4′′, E121°33′3′′). The voucher specimen used in this study was deposited in the Laboratory of Forest Silviculture and Tree Cultivation, Research Institute of Forestry, Chinese Academy of Forestry in Beijing, China (Voucher specimen: ACNIK-ZJNB2018-01). Total genome DNA was extracted by the plant genomic DNA extraction kit (DP350) (Tiangen biotech Inc., Beijing, China) and then sequenced using the Illumina Hiseq platform (Huitong biotechnology Inc., Shenzhen, China). The de novo assembly and genome annotation were respectively performed using SPAdes v3.9.0 (Bankevich et al. Citation2012) and DOGMA (Wyman et al. Citation2004). Finally, the chloroplast genome map was drawn by OGDRAW (Lohse et al. Citation2013).
Being similar with most other angiosperms, A. nikoense had a typical quadripartite structure with 155,952 bp in length, consisting of a large single-copy region (LSC: 85,720 bp), a small single-copy region (SSC: 18,072 bp), as well as a pair of inverted repeats (IRa and IRb: 26,080 bp). The total GC content of this circular DNA molecule was 37.9%. As a result of annotation, a total of 136 genes were annotated, of which 113 are unique genes, including 30 tRNAs, four rRNAs, and 79 protein-coding genes. The complete chloroplast genome characteristics of A. nikoense were similar to that of A. triflorum (Xia et al. Citation2020).
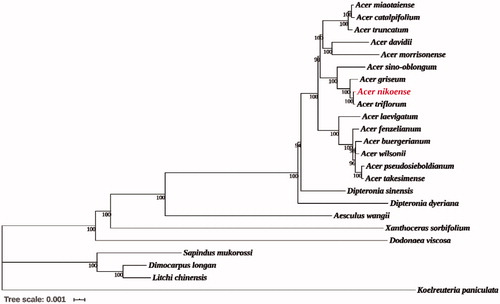
Phylogenetic relationships were reconstructed using maximum-likelihood method through MEGA v 7.0.14 (Kumar et al. Citation2016) (), based on the chloroplast genome of A. nikoense and the genomes of 23 other species downloaded from NCBI Organelle Genome Database. All sequences were aligned using MAFFT (Nakamura et al. Citation2018). The results clearly indicated that A. nikoense and A. triflorum were most closely related, followed by A. griseum, which was inconsistent with the conclusion of Wei (Citation2019), which showed the closest relationship between A. nikoense and A. griseum by SSR markers. In addition, the genus Acer had the closest relationship with Dipteronia and were clustered into monophyly by 100% bootstrap value, which has in common with the result of Xia et al. (Citation2020). Overall, the complete chloroplast genome of A. nikoense obtained in this study offered abundant genomic information for the future study on the phylogeny of Acer L.
Figure 1. Phylogenetic tree reconstruction of 24 species using maximum-likelihood (ML) based on the complete chloroplast genome sequences of A. nikoense and other 23 species. There are the bootstrap support values from 1000 replicates given at each node. Their accession numbers are as follows: Acer buergerianum: NC_034744; Acer catalpifolium: NC_041080; Acer davidii: NC_030331; Acer fenzelianum: NC_045527; Acer griseum: NC_034346; Acer pseudosieboldianum: NC_046487; Acer laevigatum: NC_042443; Acer miaotaiense: NC_030343; Acer morrisonense: NC_029371; Acer sino-oblongum: NC_040106; Acer takesimense: NC_046488; Acer triflorum: MN602455; Acer truncatum: NC_037211; Acer wilsonii: NC_040988; Aesculus wangii: NC_035955; Dimocarpus longan: NC_037447; Dipteronia dyeriana: NC_031899; Dipteronia sinensis: NC_029338; Dodonaea viscosa: NC_036099; Litchi chinensis: NC_035238; Koelreuteria paniculata: NC_037176; Sapindus mukorossi: NC_025554; Xanthoceras sorbifolium: NC_037448.
Acknowledgements
We are grateful to Zhiyong Zhu, who is a professor at Ningbo City College of Vocational Technology, for his assistance helping us collecting the leaf sample.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/, reference number MT216763.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Kim NT, Lee DS, Chowdhury A, Lee L, Cha BY, Woo JT, Woo ER, Jang JH. 2017. Acerogenin C from Acer nikoense exhibits a neuroprotective effect in mouse hippocampal HT22 cell lines through the upregulation of Nrf-2/HO-1 signaling pathways. Mol Med Rep. 16(2):1537–1543.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Kurimoto S, Sasaki YF, Suyama Y, Tanaka N, Kashiwada Y, Nakamura T. 2016. Acylated triterpene saponins from the stem bark of Acer nikoense (Aceraceae). Chem Pharm Bull. 64(7):924–929.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(Web Server issue):W575–W581.
- Nakamura T, Yamada KD, Tomii K, Katoh K. 2018. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics. 34(14):2490–2492.
- Wang S, Xie Y. 2004. China Species Red List (Vol. 1). Beijing: Higher Education Press.
- Wang WC, Chen SY, Zhang XZ. 2017. The complete chloroplast genome of the endangered Chinese paperbark maple, Acer griseum (Sapindaceae). Conserv Genet Resour. 9(4):527–529.
- Wei BY. 2019. The phylogeny study of Acer L. sect. Trifoliata Pax. Jilin, China: Northeast Normal University.
- Wu ZY, Raven PH, Hong DY. 2008. Oxalidaceae through Aceraceae. . Qian CS, Chen HY, Tang J, Wang FZ. Flora of China vol 11; Beijing: Science Press.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Xia XH, Yu XD, Fu QD, Zheng YQ, Zhang CH. 2020. Complete chloroplast genome sequence of the three-flowered maple, Acer triflorum (Sapindaceae). Mitochondrial DNA B. 5(2):1859–1860.
