Abstract
We have determined the second mitochondrial genome of Alphitobius diaperinus Panzer, 1797 collected in Gyeonggi-do, Republic of Korea. The circular mitogenome of A. diaperinus is 15,512 bp long which is slightly longer than that of the previous mitogenome of A. diaperinus. It includes 13 protein-coding genes, two ribosomal RNA genes, and 22 transfer RNAs. The base composition was AT-biased (72.4%). Intraspecific variation between two mitogenome of A. diaperinus was investigated: one SNP and one INDEL were identified, presenting the low level of intraspecific variations on mitochondrial genome.
In Korea, the lesser mealworm, Alphitobius diaperinus Panzer, 1797 (Coleoptera: Tenebrionidae), is one of the significant pests in the poultry industry (Axtell and Arends 1990). Recently, this beetle has infected in broiler chicken houses, resulting destruction of the insulate facilities of poultry houses, reduction of chick’s overall performance, and transmission of pathogenic organisms which cause substantial economic losses for poultry producers (Nguyen et al. Citation2019).
To investigate intraspecific variations of its mitochondrial genome, we completed the second mitogenome of A. diaperinus, collected in Munsan-eup, Paju-si, Gyeonggi-do, Republic of Korea (37°88′99″N, 126°77′42″E; the specimen and its DNA were deposited at the Sunchon National University, Korea, Accession number: 190925HK0010). DNA was extracted using DNeasy Blood &Tissue Kit (QIAGEN, Hilden, Germany). Raw sequences generated from Illumina HiSeqX (Macrogen, Korea) were filtered by Trimmomatic 0.33 (Bolger et al. Citation2014) and de novo assembled by Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li Citation2013), and SAMtools 1.9 (Li et al. Citation2009) under the environment of Genome Information System (GeIS; http://geis.infoboss.co.kr/). Geneious R11 11.1.5 (Biomatters Ltd, Auckland, New Zealand) was used to annotate based on the previous mitogenome of A. diaperinus (MT165524; Hong et al. Citation2020).
A. diaperinus mitogenome (GenBank accession is MT610905) is 15,512 bp long, which is slightly longer than that of the previous mitogenome of A. diaperinus. It contains 13 protein-coding genes (PCGs), 37 tRNAs, two rRNAs and its AT ratio is 72.4%. Gene order is conserved among 24 available Tenebrionidae mitogenomes.
Based on comparison of the two mitogenomes of A. diaperinus, we identified one SNP and one INDEL located in NAD4 and AT-rich regions, respectively. One SNP is non-synonymous SNP changing 142nd amino acid of NAD4 from methionine to valine. Number of intraspecific variations identified in this study is similar to that of Laodelphax striatellus between D5 and D7 isolates (Seo, Jung, et al. Citation2019) but much less than those of Hipparchia autonoe (Lee et al. Citation2020), Aphis gossypii (Park, Xi, Kim, et al. Citation2019) (Bae et al. Citationaccepted), Laodelphax striatellus (Park, Jung, et al. Citation2019; Seo, Jung, et al. Citation2019), Nilaparvata lugens (Choi et al. Citation2019; Park, Kwon, et al. Citation2019; Choi et al. Citation2020), Spodoptera frugiperda (Seo, Lee, et al. Citation2019), and Chilo suppresallis (Park, Xi, Kwon, et al. Citation2019).
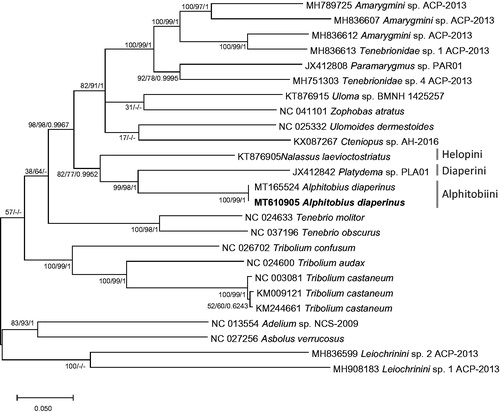
We inferred phylogenetic relationship of 25 Tenebrionidae mitogenomes, including two A. diaperinus mitogenome based on alignment of complete mito genomes by MAFFT 7.450 (Katoh and Standley Citation2013) after rearranging mitogenome sequences as same as the previous study (Hong et al. Citation2020). Bootstrapped neighbor joining, maximum likelihood, and Bayesian inference phylogenetic trees were constructed by MEGA X (Kumar et al. Citation2018) and Mr. Bayes (Huelsenbeck and Ronquist Citation2001). It displays that two mitogenomes of A. diaperinus were clustered and three neighbor tribes were also clustered in one clade as same as the previous study (Hong et al. Citation2020; ).
Figure 1. Bayesian inference (1,000,000 generations), maximum likelihood (1,000 bootstrap repeats), and neighbor joining (10,000 bootstrap repeats) phylogenetic trees of 24 Tenebrionidae mitochondrial genomes: Alphitobius diaperinus (MT610905 in this study and MT165524), Amarygmini sp. (MH789725; Partial mitochondrial genome; MH836607, and MH836612), Tenebrionidae sp. (MH836613 and MH751303), Paramarygmus sp. (JX412808; Partial mitochondrial genome), Cteniopussp. (KX087267; Partial mitochondrial genome), Zophobas atratus (NC_041101), Uloma sp. (KT876915), Ulomoides dermestoides (NC_025332), Nalassus laevioctostriatus (KT876905), Platydema sp. (JX412842), Tenebrio molitor (NC_024633), Tenebrio obscurus (NC_037196), Tribolium confusum (NC_026702), Tribolium audax (NC_024600), Tribolium castaneum (NC_003081, KM009121, and KM244661), Adelium sp. (NC_013554), Asbolus verrucosus (NC_027256), and Leiochrinini sp. (MH836599 and MH908183). Phylogenetic tree was drawn based on maximum likelihood tree. The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining phylogenetic trees and posterior probability value of Bayesian inference, respectively. Tribe names were displayed as light gray color and subfamily names were written as gray color.
Disclosure statement
The authors declare that they have no competing interests.
Data availability statement
The data that support the findings of this study are openly available in NCBI (National Center for Biotechnology Information) at https://www.ncbi.nlm.nih.gov/nuccore/MT610905.
References
- Axtell RC, Arends JJ. 1990. Ecology and management of arthropod pests of poultry. Annu Rev Entomol. 35:101–125.
- Bae Y, Park J, Iee w. (accepted). The complete mitochondrial genome of Aphis gossypii Glover, 1877 (Hemiptera: Aphididae) isolated from Plantago asiatica in Korea. DOI:10.1080/23802359.2020.1792366
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Choi NJ, Lee B-C, Park J, Park J. 2019. The complete mitochondrial genome of Nilaparvata lugens (Stål, 1854) captured in China (Hemiptera: Delphacidae): investigation of intraspecies variations between countries. Mitochondrial DNA Part B. 4(1):1677–1678.
- Choi NJ, Lee B-C, Park J, Park J. 2020. The complete mitochondrial genome of Nilaparvata lugens (Stål, 1854) captured in Guangxi province, China (Hemiptera: Delphacidae): identification of the origin of N. lugens migrated to Korea. Mitochondrial DNA Part B. 5(2):1960–1961.
- Hong K-J, Ki W, Park D-S, Yang B-K, Lee H, Park J, Lee W. 2020. The complete mitochondrial genome of Alphitobius diaperinus Panzer, 1797 (Coleoptera: Tenebrionidae. ). Mitochondrial DNA Part B. 5(3):2291–2293.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lee Y-D, Lee J, Kim D-S, Park J, Xi H, Roh J, Kim D-S, Nam SJ, Kim S-K, Song J-Y, et al. 2020. The complete mitochondrial genome of Hipparchia autonoe (Esper, 1783)(Lepidoptera: Nymphalidae): investigation of intraspecific variations on mitochondrial genome. Mitochondrial DNA Part B. 5(2):1542–1544.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Nguyen N, Yang B-K, Lee J-S, Yoon J-U, Hong K-J. 2019. Infestation status of the darkling beetle (Alphitobius diaperinus) in Broiler chicken houses of Korea. Korean J Appl Entomol. 58(3):189–196.
- Park J, Jung JK, Ho Koh Y, Park J, Seo BY. 2019. The complete mitochondrial genome of Laodelphax striatellus (Fallén, 1826)(Hemiptera: Delphacidae) collected in a mid-Western part of Korean peninsula. Mitochondrial DNA Part B. 4(2):2229–2230.
- Park J, Kwon W, Park J, Kim H-J, Lee B-C, Kim Y, Choi NJ. 2019. The complete mitochondrial genome of Nilaparvata lugens (stål, 1854) captured in Korea (Hemiptera: Delphacidae). Mitochondrial DNA Part B. 4(1):1674–1676.
- Park J, Xi H, Kim Y, Park J, Lee W. 2019. The complete mitochondrial genome of Aphis gossypii Glover, 1877 (Hemiptera: Aphididae) collected in Korean peninsula. Mitochondrial DNA Part B. 4(2):3007–3009.
- Park J, Xi H, Kwon W, Park C-G, Lee W. 2019. The complete mitochondrial genome sequence of Korean Chilo suppressalis (Walker, 1863) (Lepidoptera: Crambidae). Mitochondrial DNA Part B. 4(1):850–851.
- Seo BY, Jung JK, Ho Koh Y, Park J. 2019. The complete mitochondrial genome of Laodelphax striatellus (Fallén, 1826)(Hemiptera: Delphacidae) collected in a southern part of Korean peninsula. Mitochondrial DNA Part B. 4(2):2242–2243.
- Seo BY, Lee G-S, Park J, Xi H, Lee H, Lee J, Park J, Lee W. 2019. The complete mitochondrial genome of the fall armyworm, Spodoptera frugiperda Smith, 1797 (Lepidoptera; Noctuidae), firstly collected in Korea. Mitochondrial DNA Part B. 4(2):3918–3920.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC bioinformatics. 12(14):S2.
