Abstract
Arisaema erubescens is a well-known medicinal plant in China. In this study, we sequenced the complete chloroplast (cp) genome sequence of A. erubescens to investigate its phylogenetic relationship in the family Araceae. The cp genome was 167,607 bp in length, consisting of a pair of inverted repeats (IRa and IRb: 26,193 bp) separated by a large single-copy region (LSC: 93,660 bp) and a small single-copy region (SSC: 21,561 bp). The GC content of whole cp genome is 35.3%. De novo assembly and annotation showed the presence of 114 unique genes with 80 protein-coding genes, 30 tRNA genes, and four rRNA genes. Phylogenetic analysis indicated that A. erubescens was closely related to A. franchetianum, and the genus Arisaema was sister to the genus Pinellia.
Arisaema erubescens (Wall.) Schott is a perennial herb of the Araceae family, which is widely distributed in south China and Southeast Asian countries. It is used medically as an important antidote to treat several biological disorders (Li et al. Citation2010). With the discovery of the great medicinal value of A. erubescens and the increasing demand of the market, the wild resources of A. erubescens are decreasing nowadays. It is necessary to develop genomic resources for A. erubescens to provide intragenic information for its utilization. Chloroplast genomes are valuable sources of genetic markers for phylogenetic analyses, genetic diversity evaluation, and plant molecular identification (Dong et al. Citation2018; Sun et al. Citation2020). In this study, the complete chloroplast genome of A. erubescens was assembled to provide genomic and genetic resources for further research, and the phylogeny of family Araceae was conducted to reveal the interspecific relationships.
Fresh samples of A. erubescens were collected from Hongjiang county, Hunan province, China (27°24′38′′N, 110°25′19″E). Voucher specimen was deposited at the herbarium of Institute of Chinese Materia Medica (CMMI), China Academy of Chinese Medical Sciences with the specimen voucher number is 431281LY0254. Total genomic DNA was isolated using the CTAB method (Doyle and Doyle Citation1987). The library with 350 bp insertion size fragments was constructed and sequenced using the Illumina HiSeq platform. De novo assembly was conducted using GetOrganelle pipeline (Jin et al. Citation2018). The annotation was performed using Plann (Huang and Cronk Citation2015), with annotated complete chloroplast (cp) genome from A. ringens (NC044118) selected as reference. The annotated complete chloroplast cp genome of A. erubescens was submitted to the GenBank under the accession number MT676834.
The complete cp genome of A. erubescens was a circular DNA molecule of 167,607 bp in length, containing a large single-copy region (LSC) of 93,660 bp, a small single-copy region (SSC) of 21,561 bp, and a pair of inverted repeat (IR) regions of 26,193 bp. The overall GC-content of the cp genome was 35.3%. The IR regions had a higher GC content (41.4%) than that of the LSC (33.2%) and SSC (29.5%) regions. The cp genome contained 114 genes, including 80 protein-coding genes, 30 tRNA genes, and four rRNA genes. In these genes, six tRNA genes (trnK-UUU, trnG-UCC, trnL-UAA, trnV-UAC, trnI-GAU, and trnA-UGC) and nine protein-coding genes (atpF, ndhA, ndhB, petB, petD, rps16, rpl16, rpl2, and rpoC1) contain one intron, while two genes (ycf3 and clpP) have two introns.
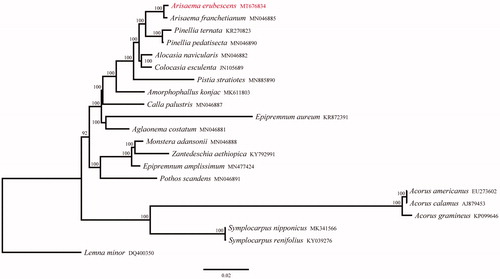
To investigate the phylogenetic relationships between A. erubescens and other related species in family Araceae, 20 complete cp genomes were downloaded from GenBank to construct a phylogenetic maximum-likelihood (ML) tree using RAxML (Stamatakis Citation2014), under the GTR + G model with 1000 rapid bootstrap replicates. Lemna minor (DQ400350) was taken as an outgroup. The phylogenetic tree showed that all 19 species forming a monophyletic group supported by a 100% bootstrap value. A. erubescens was inferred phylogenetically closest to A. franchetianum, and the genus Arisaema was sister to the genus Pinellia (). The complete cp genome of A. erubescens provided a lot of genetic information for species conservation and identification as well as the phylogenetic studies of family Araceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI https://www.ncbi.nlm.nih.gov/, reference number MT676834.
Additional information
Funding
References
- Dong W, Xu C, Wu P, Cheng T, Yu J, Zhou S, Hong DY. 2018. Resolving the systematic positions of enigmatic taxa: manipulating the chloroplast genome data of Saxifragales. Mol Phylogenet Evol. 126:321–330.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. BioRxiv. 2018:1–11.
- Li H, Zhu GH, Jin M. 2010. Arisaema. In: Wu Z, Raven P, editors. Flora of China. Beijing (China): Science Press; p. 23; St. Louis (MO): Missouri Botanical Garden Press; p. 43–69.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Sun J, Wang Y, Liu Y, Xu C, Yuan Q, Guo L, Huang L. 2020. Evolutionary and phylogenetic aspects of the chloroplast genome of Chaenomeles species. Scientific Reports.10(1):11466