Abstract
In this study, we obtained the complete mitochondrial genome of Sporobolomyces sp. using next-generation sequencing. The complete mitochondrial genome of Sporobolomyces sp. contained 15 protein-coding genes (PCG), two ribosomal RNA (rRNA) genes, and 25 transfer RNA (tRNA) genes. The total length of the Sporobolomyces sp. mitochondrial genome is 26,430 bp, and the GC content of the mitochondrial genome is 39.32%. Phylogenetic analysis based on combined mitochondrial gene dataset indicated that the mitochondrial genome of Sporobolomyces sp. exhibited a close relationship with that of Rhodotorula mucilaginosa.
The genus Sporobolomyces is a group of ballistoconidium-forming yeast. Dozens of species have been described in this genus (Satoh and Makimura Citation2008; Lorenzini et al. Citation2019). Sporobolomyces species are distributed in various habitats, such as on plant phyllodes, on the surface area of soils, and also in the fermentation system (Barahona et al. Citation2016; Arrigoni et al. Citation2018; Li, Yuan, et al. Citation2020). Limited morphological characteristics make it difficult to identify Sporobolomyces species accurately only by morphology (Hamamoto and Nakase Citation2000; Barahona et al. Citation2016; Lorenzini et al. Citation2019). Mitochondrial genome has been widely used in the phylogenetic analysis of species (Wang et al. Citation2020; Li, He, et al. Citation2020). However, up to now, no mitochondrial genome from the genus Sporobolomyces has been reported. The mitochondrial genome of Sporobolomyces sp. will promote the understanding of the phylogeny, evolution, and taxonomy of this important yeast.
The specimen (Sporobolomyces sp.) was collected from Jilin, China (129.32 E; 42.53 N). The specimen was stored in the Culture Collection Center of Chengdu University (No. Spo_ns1). The complete mitochondrial genome of Sporobolomyces sp. was sequenced and assembled according to previously described methods (Li, Liao, et al. Citation2018; Li, Xiang, et al. Citation2019; Wang et al. Citation2020). Briefly, the total genomic DNA of Sporobolomyces sp. was extracted using a Fungal DNA Kit D3390-00 (Omega Bio-Tek, Norcross, GA, USA). The extracted genomic DNA was purified using a Gel Extraction Kit (Omega Bio-Tek, Norcross, GA, USA). The purified DNA was stored in Chengdu University (No. DNA_ Spo_ns1). We constructed sequencing libraries with the purified genomic DNA using a NEBNext® Ultra™ II DNA Library Prep Kit (NEB, Beijing, China). Whole genomic sequencing (WGS) of Sporobolomyces sp. was conducted using the Illumina HiSeq 2500 Platform (Illumina, SanDiego, CA). We de novo assembled the mitochondrial genome of Sporobolomyces sp. using SPAdes 3.9.0 (Bankevich et al. Citation2012; Li, Ren, et al. Citation2020). The complete mitochondrial genome of Sporobolomyces sp. was annotated according to previously described methods (Li, Chen, et al. Citation2018; Li, Wang, et al. Citation2018).
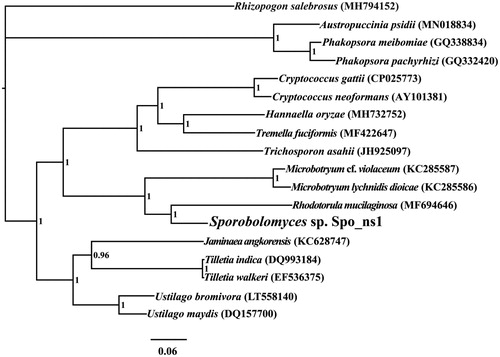
We found the complete mitochondrial genome of Sporobolomyces sp. is 26,430 bp in length, with the base composition as follows: A (30.82%), T (29.84%), G (20.35%), and C (18.97%). The complete mitochondrial genome of Sporobolomyces sp. contains 15 protein-coding genes, two ribosomal RNA genes (rns and rnl), and 25 transfer RNA (tRNA) genes. To investigate the phylogenetic positions of Sporobolomyces sp., we constructed a phylogenetic tree for 18 Basidiomycota species. Rhizopogon salebrosus from the Boletales order was set as the outgroup (Li, Ren, et al. Citation2019). The phylogenetic tree was constructed using the Bayesian analysis (BI) method based on the combined 14 core protein-coding genes according to previously described methods (Li, Wang, Jin, Chen, Xiong, Li, Liu, et al. Citation2019; Li, Wang, Jin, Chen, Xiong, Li, Zhao, et al. Citation2019; Li, Yang, et al. Citation2020). As shown in the phylogenetic tree (), the mitochondrial genome of Sporobolomyces sp. exhibited a close relationship with that of Rhodotorula mucilaginosa (Gan et al. Citation2017).
Disclosure statement
The authors have declared that no competing interests exist.
Data availability statement
This mitogenome of Sporobolomyces sp. was submitted to GenBank under the accession number of MT663151. (https://www.ncbi.nlm.nih.gov/nuccore/MT663151).
Additional information
Funding
References
- Arrigoni E, Antonielli L, Pindo M, Pertot I, Perazzolli M. 2018. Tissue age and plant genotype affect the microbiota of apple and pear bark. Microbiol Res. 211:57–68.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Barahona S, Yuivar Y, Socias G, Alcaino J, Cifuentes V, Baeza M. 2016. Identification and characterization of yeasts isolated from sedimentary rocks of Union Glacier at the Antarctica. Extremophiles. 20(4):479–491.
- Gan HM, Thomas BN, Cavanaugh NT, Morales GH, Mayers AN, Savka MA, Hudson AO. 2017. Whole genome sequencing of Rhodotorula mucilaginosa isolated from the chewing stick (Distemonanthus benthamianus): insights into Rhodotorula phylogeny, mitogenome dynamics and carotenoid biosynthesis. PeerJ. 5:e4030.
- Hamamoto M, Nakase T. 2000. Phylogenetic analysis of the ballistoconidium-forming yeast genus Sporobolomyces based on 18S rDNA sequences. Int J Syst Evol Microbiol. 50 Pt 3:1373–1380.
- Li Q, Chen C, Xiong C, Jin X, Chen Z, Huang W. 2018. Comparative mitogenomics reveals large-scale gene rearrangements in the mitochondrial genome of two Pleurotus species. Appl Microbiol Biotechnol. 102(14):6143–6153.
- Li Q, He X, Ren Y, Xiong C, Jin X, Peng L, Huang W. 2020. Comparative mitogenome analysis reveals mitochondrial genome differentiation in ectomycorrhizal and asymbiotic Amanita species. Front Microbiol. 11:1382.
- Li Q, Liao M, Yang M, Xiong C, Jin X, Chen Z, Huang W. 2018. Characterization of the mitochondrial genomes of three species in the ectomycorrhizal genus Cantharellus and phylogeny of Agaricomycetes. Int J Biol Macromol. 118:756–769.
- Li Q, Ren Y, Shi X, Peng L, Zhao J, Song Y, Zhao G. 2019. Comparative mitochondrial genome analysis of two ectomycorrhizal fungi (Rhizopogon) reveals dynamic changes of intron and phylogenetic relationships of the subphylum Agaricomycotina. IJMS. 20(20):5167.
- Li Q, Ren Y, Xiang D, Shi X, Zhao J, Peng L, Zhao G. 2020. Comparative mitogenome analysis of two ectomycorrhizal fungi (Paxillus) reveals gene rearrangement, intron dynamics, and phylogeny of basidiomycetes. IMA Fungus. 11(1):12.
- Li Q, Wang Q, Chen C, Jin X, Chen Z, Xiong C, Li P, Zhao J, Huang W. 2018. Characterization and comparative mitogenomic analysis of six newly sequenced mitochondrial genomes from ectomycorrhizal fungi (Russula) and phylogenetic analysis of the Agaricomycetes. Int J Biol Macromol . 119:792–802.
- Li Q, Wang Q, Jin X, Chen Z, Xiong C, Li P, Liu Q, Huang W. 2019. Characterization and comparative analysis of six complete mitochondrial genomes from ectomycorrhizal fungi of the Lactarius genus and phylogenetic analysis of the Agaricomycetes. Int J Biol Macromol. 121:249–260.
- Li Q, Wang Q, Jin X, Chen Z, Xiong C, Li P, Zhao J, Huang W. 2019. Characterization and comparison of the mitochondrial genomes from two Lyophyllum fungal species and insights into phylogeny of Agaricomycetes. Int J Biol Macromol. 121:364–372.
- Li Q, Xiang D, Wan Y, Wu Q, Wu X, Ma C, Song Y, Zhao G, Huang W. 2019. The complete mitochondrial genomes of five important medicinal Ganoderma species: features, evolution, and phylogeny. Int J Biol Macromol. 139:397–408.
- Li Q, Yang L, Xiang D, Wan Y, Wu Q, Huang W, Zhao G. 2020. The complete mitochondrial genomes of two model ectomycorrhizal fungi (Laccaria): features, intron dynamics and phylogenetic implications. Int J Biol Macromol. 145:974–984.
- Li AH, Yuan FX, Groenewald M, Bensch K, Yurkov AM, Li K, Han PJ, Guo LD, Aime MC, Sampaio JP, Jindamorakot S, et al. 2020. Diversity and phylogeny of basidiomycetous yeasts from plant leaves and soil: proposal of two new orders, three new families, eight new genera and one hundred and seven new species. Stud Mycol. 96:17–140.
- Lorenzini M, Zapparoli G, Azzolini M, Carvalho C, Sampaio JP. 2019. Sporobolomyces agrorum sp. nov. and Sporobolomyces sucorum sp. nov., two novel basidiomycetous yeast species isolated from grape and apple must in Italy. Int J Syst Evol Microbiol. 69(11):3385–3391.
- Satoh K, Makimura K. 2008. Sporobolomyces koalae sp. nov., a basidiomycetous yeast isolated from nasal smears of Queensland koalas kept in a Japanese zoological park. Int J Syst Evol Microbiol. 58(Pt 12):2983–2986.
- Wang X, Song A, Wang F, Chen M, Li X, Li Q, Liu N. 2020. The 206 kbp mitochondrial genome of Phanerochaete carnosa reveals dynamics of introns, accumulation of repeat sequences and plasmid-derived genes. Int J Biol Macromol. 162:209–219.