Abstract
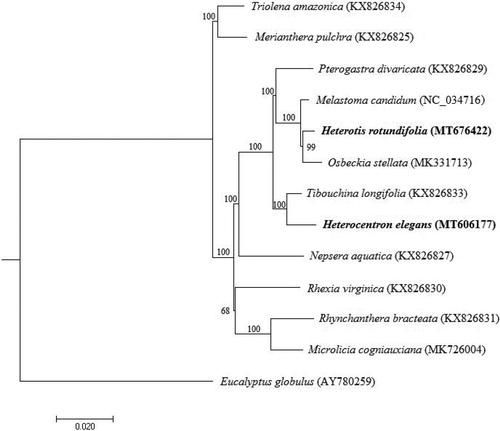
Heterotis and Heterocentron are two shrub genera in the tribe Melastomateae (Melastomataceae). Heterotis rotundifolia (Hr) and Heterocentron elegans (He) are both widely used as ornamental plants in the tropical regions. In this study, the complete chloroplast (cp) genomes of Hr and He were sequenced, assembled, and annotated. The cp genome sizes of Hr and He are 156,336 bp and 156,514 bp, respectively. The cp genome of Hr contains a total of 132 genes, including 87 protein-coding, 37 tRNA, and eight rRNA genes, while that of He has 131 genes, including 87 protein-coding, 36 tRNA, and eight rRNA genes. Phylogenetic analysis reveals that Hr is sister to Osbeckia stellata, while He is sister to Tibouchina longifolia.
The tribe Melastomateae (Melastomataceae) is composed of about 12 genera, including Heterotis Benth and Heterocentron Hook & Arn (Veranso-Libalah et al. Citation2017). Heterotis, comprising about 10 species (Plant List Citation2013), is natively distributed in tropical Africa (Singh and Ranjan Citation2015). Heterocentron, containing about 29 species, is native to Mexico and Central America (Almeda Citation1999). Heterotis rotundifolia (Hr) and Heterocentron elegans (He) are both used for ornamental purposes and have been introduced to tropical regions in the world. Herein, we report the complete chloroplast (cp) genome sequences of these two species as a genetic resource for future molecular phylogeny studies of the tribe Melastomateae.
Genomic DNAs were extracted from fresh leaf samples of Hr and He, which are collected from South China Botanical Graden, Guangzhou, Guangdong, China (23°10′59″N, 113°22′01″E), and the voucher specimens (Hr: no. ZXJ-1902; He: no. ZXJ-2001) are deposited at Herbarium of Sun Yat-sen University (SYS), Guangzhou, China. Shotgun DNA libraries were constructed and then sequenced on an Illumina HiSeq X Ten platform. About 5 Gb paired-end sequence data were generated for each species and fed into the NOVOPlasty pipeline (Dierckxsens et al. Citation2017) for the cp genome assembly. Gene annotation was conducted using GeSeq (Tillich et al. Citation2017) and then manually corrected when necessary.
The complete cp genome sizes of Hr and He are 156,336 bp and 156,514 bp, respectively. They both have typical quadripartite structure. The cp genome of Hr possesses a total of 132 genes, including 87 protein-coding, 37 transfer RNA (tRNA), and eight ribosomal RNA (rRNA) genes. The cp genome of He contains 131 genes, including 87 protein-coding, 36 tRNA, and eight rRNA genes.
Phylogenetic analysis was performed using RAxML (Stamatakis Citation2014), based on the complete cp genome sequences of 13 taxa representing 12 genera from Melastomataceae and an outgroup species, Eucalyptus globulus, from Myrtaceae (). The maximum-likelihood tree reveals that both species are nested within the tribe Melastomateae, among which Hr is sister to Osbeckia stellata, while He is most closely related to Tibouchina longifolia.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the NCBI GenBank (http://www.ncbi.nlm.nih.gov) under accession number MT676422 and MT606177.
Additional information
Funding
References
- Almeda F. 1999. Heterocentron evansii (Melastomataceae): a new species from Pico Bonito National Park. Novon. 9(2):127–130.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Singh LJ, Ranjan V. 2015. Heterotis Benth. (Melastomataceae): a new addition to Indian flora from Andaman Islands. Geophytology. 45(1):101–106.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Plant List. 2013. Version 1.1. http://www.theplantlist.org/.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Veranso-Libalah MC, Stone RD, Fongod AGN, Couvreur TLP, Kadereit G. 2017. Phylogeny and systematics of African melastomateae (Melastomataceae). Taxon. 66(3):584–614.