Abstract
Carya ovata is a slow-growing, long-lived deciduous species that belongs to section Carya of genus Carya. In this study, we de novo assembled the complete chloroplast genome of C. ovata, and analyzed its phylogenetic relationship. The circular genome was 160,765 bp in length, comprising a large single-copy region (89,975 bp), a small single-copy region (18,788 bp), and a pair of inverted repeat regions (26,001 bp each). The chloroplast genome was predicted to contain 131 genes, including 83 protein-coding genes, 40 transfer RNA (tRNA) genes, and 8 ribosomal RNA (rRNA) genes. Overall, the GC content of the chloroplast genome was 36.16%. Phylogenetic analysis suggested that C. ovata was closely related to C. illinoinensis, a representative of section Apocarya within the genus Carya.
Carya ovata, commonly known as shagbark hickory, is a member of family Juglandaceae (Thompson and Grauke Citation1991). This species is distinguished by its conspicuous and persistent dark outer bud scales over the winter buds. Previously, the phylogenetic position of C. ovata was mainly determined according to its geographical distribution and morphology (Manos and Stone Citation2001). An application of comparative genomic date to illustrate its phylogenetic relationships is still lacking. Chloroplast genome of angiosperms is maternal inheritance and displays a relatively slow rate of evolution compared to nuclear genomes (Daniell et al. Citation2016; Liu et al. Citation2016; Mo et al. Citation2020). Therefore, the polymorphic regions in chloroplast genomes are commonly used for phylogenetic analysis (Chen et al. Citation2018). In this study, we de no assembled the C. ovata chloroplast genome and analyzed its phylogenetic position on the molecular level.
Fresh and healthy leaves of C. ovata was collected from experimental farm of Jiangsu Academy of Forestry (Nanjing, China 118°45′57.30″E, 31°51′27.94″N), and stored at −80 °C until further use. Its voucher specimens are preserved in the Herbarium of Jiangsu Academy Forestry (JAF: Lyu20200512-3). Total genomic DNA of C. ovata was isolated by using Tiangen Plant Genomic DNA Kit (Tiangen Biotech Co., Beijing, China). The extracted DNA was broken into small fragments to be used for paired end library construction. The genomic library was sequenced on an Illumina Hiseq 4000 platform. After the completion of sequencing, raw reads were generated and were further processed to obtain clean reads by using the Trimmomatic (v0.32) software. The annotated chloroplast genome of C. illinoinensis was downloaded from GenBank (accession number: MH909600), and was used as reference for C. ovata chloroplast genome assembly through the NOVOPlasty software. Chloroplast genome annotation was performed by using the online annotation program DOGMA, and manually checked by Blast search. The annotated chloroplast genome was deposited in GenBank (Accession number: MT701613).
The C. ovata chloroplast genome was 160,765 bp in length, including two copies of inverted repeats (IRa and IRb) of 26,001 bp that were separated by a large single-copy (LSC) region of 89,975 bp and a small single-copy (SSC) region of 18,788 bp. The C. ovata chloroplast genome was predicted to contain 131 genes: 83 protein-coding genes, 40 tRNA genes, and 8 rRNA genes, 18 of which were duplicated in the IR regions, giving 113 unique genes totally. The chloroplast genome consisted of 48.76% coding regions and 51.24% non-coding regions (including both intergenic spaces and introns). Overall, the GC content contained in the chloroplast genome is 36.16%.
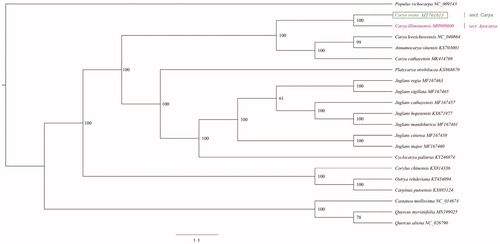
Phylogenetic analysis was performed based on the complete chloroplast genome sequences of 21 species: 14 from Juglandaceae, 3 from Betulaceae, 3 from fagaceae, and 1 outgroup (Populus richocarpa). We firstly used MAFFT to align the multiple sequences, and then applied IQ-tree to construct phylogenetic tree with the maximum-likelihood method (Katoh and Standley Citation2013; Nguyen et al. Citation2015) (). The genus Carya is currently divided into three section: sect. Carya, sect. Apocarya, and sect. Sinocarya (Manos and Stone Citation2001). Carya ovata and C. illinoinensis are representatives of sect. Carya and sect. Apocarya, respectively. Carya cathayensis and C. kweichowensis are the representative species of sect. Sinocarya. Our phylogenetic analysis indicated that C. ovata was closely related to the species of sect. Apocarya, represented by C. illinoinensis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at http://www.ncbi.nlm.nih.gov/, reference number MT701613.
Additional information
Funding
References
- Chen S, Xu Z, Zhao Y, Zhong X, Li C, Yang G. 2018. Structural characteristic and phylogenetic analysis of the complete chloroplast genome of Dianthus caryophyllus. Mitochondrial DNA Part B. 3(2):1131–1132.
- Daniell H, Lin CS, Yu M, Chang WJ. 2016. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17(1):134.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Liu YC, Lin BY, Lin JY, Wu WL, Chang CC. 2016. Evaluation of chloroplast DNA markers for intraspecific identification of Phalaenopsis equestris cultivars. Sci Hortic. 203:86–94.
- Manos PS, Stone DE. 2001. Evolution, phylogeny, and systematics of the Juglandaceae. Ann Missouri Botanical Garden. 88(2):231–269.
- Mo Z, Lou W, Chen Y, Jia X, Zhai M, Guo Z, Xuan J. 2020. The chloroplast genome of Carya illinoinensis: genome structure, adaptive evolution, and phylogenetic analysis. Forests. 11(2):207.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Thompson TE, Grauke LJ. 1991. Pecans and other hickories (Carya). Acta Hortic. 290(290):839–906.