Abstract
Ulmus parvifolia is a promising tree species for landscaping. In this study, the complete genome of U. parvifolia was reported using next-generation sequencing technology. The chloroplast genome was a circular double-stranded DNA molecule with 159,182 bp in length. It contained a large single copy (LSC) region of 87,838 bp, a small single copy (SSC) region of 18,750 bp, and two inverted repeat (IRa and IRb) regions of 26,297 bp each, which exhibited a typical quadripartite structure. A total of 133 genes were identified, including 84 protein-coding genes, 41 tRNA genes, and eight rRNA genes. The overall GC content in the chloroplast genome was 35.59%. Phylogenetic analysis indicated that U. parvifolia, as a representative of Sect. Microptelea within the Ulmus genus, is sister to the species of Sect. Ulmus.
Ulmus parvifolia, the alternate name Chinese elm, is a promising landscape species in family Ulmaceae with a wide geographic distribution. It exhibits outstanding adaptability to a diverse range of adverse environmental conditions. It has been proven to be highly resistant to elm leaf beetle and Dutch elm disease (Bosu and Wagner Citation2007). In the meantime, U. parvifolia is recognized as having drought, heat, and cold resistance traits (Lyu et al. Citation2020). Nowadays, it is commonly planted on lawns, along streets, and in parks, which is ecologically important (Thakur and Karnosky Citation2007). Nevertheless, very limited genomic data is available on this species. Chloroplast genome is characterized by small genome size, maternal transmission, and low mutation rate, which make the chloroplast-derived markers suitable for species identification, phylogenetic analysis, and population genetics studies (Li et al. Citation2019; Cai et al. Citation2020; Mo et al. Citation2020). In this study, the complete chloroplast genome of U. parvifolia was de novo assembled, annotated, and analyzed phylogenetically.
Fresh leaves were collected from experimental farm of Jiangsu Academy of Forestry (Nanjing, China 118°45′57.30″E, 31°51′27.94″N) and were deposited in the Herbarium of Jiangsu Academy Forestry (JAF: Lyu20200512-3). Genomic DNA was extracted using Universal Plant Total DNA Extraction Kit (BioTeke, Beijing, China) according to the manufacturer's instruction. A paired-end library with approximate insert lengths of 300 bp was constructed by an Illumina Hiseq library kit (Illumina, San Diego, USA), and then the paired reads were sequenced on the Illumina Hiseq 4000 platform. De novo assembly and annotation of the chloroplast genome were conducted using NOVOPlasty and DOGMA, respectively.
The length of the entire chloroplast genome of was 159,182 bp. Its genome has a conserved quadripartite structure, consisting of 87,838 bp large single copy (LSC) region, 18,750 bp small single copy (SSC) region, and two 26,297 bp inverted repeat (IRa and IRb) regions. The chloroplast genome was predicted to make up of 133 genes: 84 protein-coding genes, 41 tRNA genes, and eight rRNA genes, of which, seven protein-coding genes, nine tRNA genes, and four rRNA genes were duplicated in the IR regions. The containing GC content in the chloroplast genome of U. parvifolia was 35.59%, and the corresponding values for the LSC, SSC, and IR regions were 33.05%, 28.56%, and 42.35%, respectively.
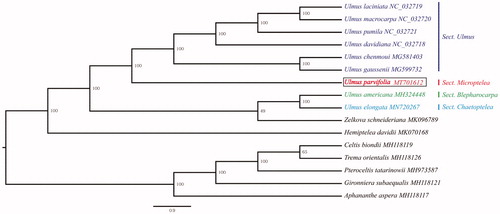
To determine the phylogenetic status of U. parvifolia, a phylogenetic tree was built using IQ-tree software (Nguyen et al. Citation2015) with the maximum-likelihood algorithm based on the chloroplast genome sequences of 16 representatives of Ulmaceae. Complete chloroplast genome sequence alignment was carried out using MAFFT program (Katoh and Standley Citation2013). The phylogenetic analysis showed that the seven species of Ulmus genus formed a paraphyletic clade with high support (bootstrap value 100%). Based on sectional-level taxonomic system, the genus Ulmus could be divided into four sections: Sect. Ulmus, Sect. Microptelea, Sect. Blepharocarpa, Sect. Trichoptelea and Sect. Chaetoptelea (Zhang et al. Citation2018). Ulmus parvifolia is a representatives of Sect. Microptelea, and our analysis revealed that it is sister to the species of Sect. Ulmus within the Ulmus genus. The chloroplast genomic resources reported here will provide a basis for future studying the phylogeny, genetic diversity, and adaptation of U. parvifolia ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at http://www.ncbi.nlm.nih.gov/, reference number MT701612.
Additional information
Funding
References
- Bosu PP, Wagner MR. 2007. Effects of induced water stress on leaf trichome density and foliar nutrients of three elm (Ulmus) species: implications for resistance to the elm leaf beetle. Environ Entomol. 36 (3):595–601.
- Cai Y, Tarin MWK, Fan L, He T, Xie D, Rong J, Zheng Y. 2020. Complete chloroplast genome of Morinda parvifolia (Rubiaceae), a traditional medicinal plant in China. Mitochondrial DNA B. 5 (2):1845–1847.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30 (4):772–780.
- Li DM, Zhao CY, Zhu GF, Xu YC. 2019. Complete chloroplast genome sequence of Amomum villosum. Mitochondrial DNA B. 4 (2):2673–2674.
- Lyu Y, Dong X, Huang L, Zheng J, He X, Sun H, Jiang Z. 2020. SLAF-seq uncovers the genetic diversity and adaptation of Chinese elm (Ulmus parvifolia) in eastern China. Forests. 11 (1):80.
- Mo Z, Lou W, Chen Y, Jia X, Zhai M, Guo Z, Xuan J. 2020. The chloroplast genome of Carya illinoinensis: genome structure, adaptive evolution, and phylogenetic analysis. Forests. 11 (2):207.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32 (1):268–274.
- Thakur R, Karnosky D. 2007. Micropropagation and germplasm conservation of Central Park Splendor Chinese elm (Ulmus parvifolia Jacq. 'A/Ross Central Park') trees. Plant Cell Rep. 26 (8):1171–1177.
- Zhang QY, Huang J, Jia LB, Su T, Zhou ZK, Xing YW. 2018. Miocene Ulmus fossil fruits from Southwest China and their evolutionary and biogeographic implications. Rev Palaeobot Palynol. 259:198–206.