Abstract
The complete plastome of Rhoiptelea chiliantha was sequenced and assembled in this study. The circular plastome of R. chiliantha is 161,702 bp in size, which contains a large single-copy region (LSC, 90,447 bp), a short single-copy region (SSC, 19,081 bp) and two inverted repeat sequences (IRs, 26,087 bp, each). It is totally comprised of 133 genes, including 88 protein-coding genes, eight ribosomal RNA, and 37 transfer RNA. The phylogenetic analysis shows that R. chiliantha is sister to the clade including remaining Juglandaceae species.
Rhoiptelea chiliantha, a deciduous tree commonly known as the horsetail tree, belongs to the family Juglandaceae (Angiosperm Phylogeny Group Citation2016). The species is endemic to southwest China and north Vietnam. Its high quality wood has been widely used in building and to make furniture and utensils (Fu et al. Citation2003). With the increase of demand, R. chiliantha resource has been greatly damaged. It has been included in the Chinese Species Red List (Wang and Xie Citation2004) and is also listed as a vulnerable species in the IUCN Red List (World Conservation Monitoring Centre Citation1998). To promote the conservation of this species, a comprehensive genomic resource is necessary to be conducted. In the present research, we sequenced and assembled the complete plastome of R. chiliantha using high throughput Illumina sequencing technology.
Samples of R. chiliantha were collected from the Botanical Garden of Kunming Institute of Botany, Chinese Academy of Sciences (25°8′21′′N, 102°44′25′′E). Voucher specimen (Y. Ji 2017129) was deposited in the Herbarium of Kunming Institute of Botany, Chinese Academy of Sciences (KUN). Genomic DNA was isolated from ∼20 mg silica gel dried leaf tissues using the CTAB method (Doyle and Doyle Citation1987). Genomic DNA was fragmented into 500 bp to construct a pair-end library. Illumina libraries were prepared according to manufacturer's protocol (Illumina, San Diego, CA), and then sequenced on the Illumina HiSeq 2000 system. The Illumina raw data were used to assemble the complete plastome (Jin et al. Citation2018), using the plastome of Carya illinoinensis (GenBank Accession No. MH188302) as reference. The assembled genome was annotated in Geneious V10.2 (Kearse et al. Citation2012), and manually checked for start and stop codons and intron/exon boundaries. The plastome sequence of R. chiliantha was deposited in the NCBI GenBank database under the accession number MT701585.
The circular plastome of R. chiliantha is 161,702 bp in size, which contains a large single-copy region (LSC, 90,447 bp), a short single-copy region (SSC, 19,081 bp) and two inverted repeat sequences (IRs, 26,087 bp, each). It is totally comprised of 133 genes (115 unique genes), including 88 protein-coding genes (PCGs, 81 unique genes), eight ribosomal RNA (rRNAs, four unique genes), and 37 transfer RNA (tRNAs, 30 unique genes). The overall G/C content in the R. chiliantha plastome is 36.10%, and the corresponding value for LSC, SSC, and IR region are 33.60, 29.80, and 42.50%, respectively.
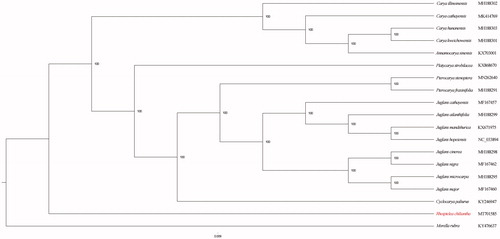
To identify the phylogenetic position of R. chiliantha, we constructed the maximum likelihood (ML) (Stamatakis Citation2014) phylogenetic tree using species within the Family Juglandaceae. Myrica rubra (GenBank Accession No. KY476637) was used to root the tree. ML analysis was conducted with 1000 bootstraps under the GTRCAT substitution model. As a result, the topology of the phylogenetic tree shows that R. chiliantha is sister to the clade including remaining Juglandaceae species ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov, reference number MT701585.
Additional information
Funding
References
- Angiosperm Phylogeny Group. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linnean Soc. 181(1):1–20.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Fu LG, Xin YQ, Bruce B. 2003. Rhoipteleaceae. In: Wu ZY, Raven PH, editors. Flora of China. Vol. 5. Beijing and St. Louis: Science Press and Missouri Botanical Garden Press; p. 20.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv.doi:10.1101/256479.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Stamatakis A. 2014. Raxml version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang S, Xie Y. 2004. Chinese species red list. Beijing: Higher Education Press.
- World Conservation Monitoring Centre. 1998. Rhoiptelea chiliantha. The IUCN Red List of Threatened Species.