Abstract
Cletus rubidiventris is a crop pest, especially for rice. This study first reported the complete mitochondrial genome of this species. The total length of mitochondrial genome is 15,590 bp and including 13 PCGs, 22 tRNA genes, and 2 rRNA genes, with 31.8% T, 15.8% C, 41.6% A, and 10.8% G. The overall GC content of the genome is 27%. The mitochondrial genome order, nucleotide composition, and codon usage pattern is similar to C. punctiger. The phylogenetic tree shows that C. rubidiventris belong to the Coreidae.
Cletus rubidiventris, belonging to the Coreidae, is a pest of rice and feeding on young panicle. It often causes a decline of rice yield. So far, the molecular studies on the mitochondrial total sequence of the genus Cletus have only completed the determination of mtDNA sequence in C. punctiger (Zhang et al. Citation2019). Herein, the complete mitochondrial genome of C. rubidiventris, its genetic structures and the phylogenetic relationship were first discussed.
The specimens of C. rubidiventris were collected from campus of Guizhou Normal University (26°22′50.30″N, 106°38′11.72″E) and stored at Chengdu Research Base of Giant Panda Breeding (voucher number: CPC201913). The complete mitochondrial genome was sequenced using Illumina HiseqXten and was assembled using SOAPdenovo2 (Luo et al. Citation2012). Mictis tenebrosa (Accession number: NC_042811) was used as reference genome (Liu et al. Citation2019). The mitogenome sequence was aligned and analyzed by MEGA 5.0 (Tamura et al. Citation2011). Phylogenetic analysis by maximum likelihood (ML) was performed using RAxML 8.2.10 (Stamatakis Citation2014) under the GTRGAMMA model, using 1000 bootstrap replicates.
The complete mitochondrial genome of C. rubidiventris is 15,590 bp in length, it is contained 37 typical mitochondrial genes (13 PCGs, 22 tRNAs, and 2 rRNAs). The total base composition of the mitochondrial genome is 31.8% T, 15.8% C, 41.6% A, and 10.8% G, and with 27% GC content, which is similar to C. punctiger. Five PCGs (COII, ND1, ND2, ND3 and ND4L) starting with ATT initiation codon, three PCGs (COIII, ND4, ND5 and ATP6) starting with the ATG codon, two PCGs (CYTB and ND6) with ATA codon, COI and ATP8 with TTG and ATC codon, respectively. Nine PCGs used TAA as stop codon, ND4 and ND5 used TAG as stop codon, but CYTB and ND1 used TA- as in complete stop codon. Among the 13 protein-coding genes, the longest gene was ND5 (1713 bp), while the shortest gene was ATP8 (160 bp).
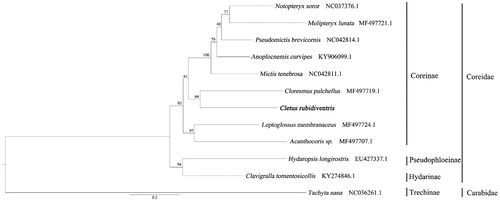
The ML phylogenetic tree was constructed with the whole mitochondrial genome of Coreidae and set Tachyta nana as outgroup (). Nodes with >70 bootstrap (BS; Hillis and Bull Citation1993) were considered to the strongly supported. The result of phylogeny tree shows C. rubidiventris nested with Coreidae and it is sister to Cloresmus pulchellus with highly supports (BS = 99). This is the first time to reveal the complete mitochondrial genome of C. rubidiventris. This not only can provide important data for the phylogenetic evolution research and genetic identification, but also contribute to the conservation of ecology.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT702884.
Additional information
Funding
References
- Hillis DM, Bull JJ. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 42(2):182–192.
- Liu YQ, Li H, Song F, Zhao YS, Wilson JJ, Cai WZ. 2019. Higher-level phylogeny and evolutionary history of Pentatomomorpha (Hemiptera: Heteroptera) inferred from mitochondrial genome sequences. Syst Entomol. 44(4):810–819.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Giga Sci. 1(1):18.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28(10):2731–2739.
- Zhang HY, Yu XL, Jiang WL, Gao H, Tan W, Wang WX, Liu YX, Tian XX. 2019. The complete mitochondrial genome sequence of Cletus punctiger (Heteroptera: Coreidae). Mitochondrial DNA Part B. 4(2):3421–3422.