Abstract
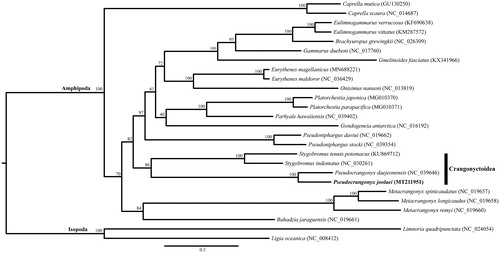
The complete mitochondrial genome of a subterranean pseudocrangonyctid amphipod, Pseudocrangonyx joolaei, was determined in this paper. The complete mitogenome of P. joolaei was 14,814 bp in length with the typical 13 protein-coding genes, 22 transfer RNAs, two ribosomal RNAs, and a control region (CR). The gene order of P. joolaei was unique in that the CR was an inversion, and the gene order of Pseudocrangonyx was not concordant when compared to that of P. daejeonensis, a subterranean amphipod found in Korea. A maximum-likelihood tree, constructed based on 26 eumalacostracan mitogenomes, confirmed that P. joolaei supported monophyly in the family Pseudocrangonyctidae and is most closely related to the superfamily Crangonyctoidea.
Amphipods are the most diverse group in groundwater communities, and the restricted habitat and distribution of stygobitic amphipods are important for the study of biogeography (Holsinger Citation1994). The genus Pseudocrangonyx, belonging to the superfamily Crangonyctoidea, is an important component of Holarctic subterranean habitats (Holsinger Citation1993, Citation1994), and is the most diverse taxon among the subterranean-amphipod genera found in East Asia, i.e., the Korean Peninsula, Japan, Eastern China, and the Russian Far East (Sidorov and Holsinger Citation2007; Tomikawa and Nakano Citation2018). However, the evolutionary history of Pseudocrangonyx is poorly studied, with most studies focusing on species revision and discovery (Zhao and Hou Citation2017). To date, the Pseudocrangonyx mitogenome has been fully determined only in P. daejeonensis, and the gene arrangement of P. daejeonensis shows many differences with respect to the typical pan-crustacean ground pattern (Lee et al. Citation2018). Moreover, the gene arrangement in the newly determined P. joolaei was inconsistent from that of P. daejeonensis despite both belonging to the same genus. Previous studies on subterranean amphipods inhabiting Europe and North America have highlighted the variability of the gene order in their mitogenome (Bauzà-Ribot et al. Citation2009; Aunins et al. Citation2016). Therefore, it remains unknown whether the mitogenome order among Pseudocrangonyx amphipods is variable. Accordingly, additional knowledge regarding the mitogenome of another Pseudocrangonyx species will lead us to a better understanding of the evolutionary history of gene arrangement in stygobitic amphipods.
Individuals of Pseudocrangonyx were collected from cave groundwater in Korea (36°47.13′N, 127°57.76′E). Mitochondrial DNA extraction, sequencing, and gene annotation were performed using the methods described by Song et al. (Citation2016). The extracted mitochondrial DNA has been deposited in the DNA collection at the National Institute of Biological Resources, Incheon, South Korea (deposit no. NIBRGR0000619445). A maximum-likelihood tree was constructed using IQ-tree 1.6.3 with mtZOA + F+R6 model (Nguyen et al. Citation2015; Chernomor et al. Citation2016), based on the concatenated sequences of 10 protein-coding genes (ATP6, COX1, COX2, COX3, CYTB, NAD1, NAD2, NAD3, NAD4, and NAD5) from 26 eumalacostracan species, including the present sequence and two isopods as outgroup taxa. The cox1 sequence of the extracted DNA was concordant with that from the paratype of P. joolaei (NCBI accession no. LC467002; Lee et al. Citation2020), and thus, the taxonomic identity of the present material was unquestionably clarified.
The complete mitogenome of P. joolaei (NCBI accession no. MT211951) was 14,814 bp in length and contained 13 protein-coding genes, 22 transfer RNAs, two ribosomal RNAs, and a control region. The gene arrangement of the complete mitogenome of P. joolaei was almost identical to the typical pan-crustacean ground pattern except the location of a control region. The control region of P. joolaei was found to be located between NAD1 and 16S rRNA. According to the gene arrangements of the amphipod revealed so far, the translocation of the control region is found only in Pseudocrangonyx; the exception being two lysianassoidean species inhabiting the deep sea, Eurythenes magellanicus and E. maldoror (Li et al. Citation2020).
The obtained maximum-likelihood tree showed monophyly of the family Pseudocrangonyctidae and this lineage was most closely related to a clade consisting of Stygobromus amphipods belonging to the superfamily Crangonyctoidea ().
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov, reference number MT211951.
Additional information
Funding
References
- Aunins AW, Nelms DL, Hobson CS, King TL. 2016. Comparative mitogenomic analyses of three North American stygobiont amphipods of the genus Stygobromus (Crustacea: Amphipoda). Mitochondrial DNA Part B. 1(1):560–563.
- Bauzà-Ribot MM, Jaume D, Juan C, Pons J. 2009. The complete mitochondrial genome of the subterranean crustacean Metacrangonyx longipes (Amphipoda): a unique gene order and extremely short control region. Mitochondrial DNA. 20(4):88–99.
- Chernomor O, von Haeseler A, Minh BQ. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol. 65(6):997–1008.
- Holsinger JR. 1993. Biodiversity of subterranean amphipod crustaceans: global patterns and zoogeographic implications. J Nat Hist. 27(4):821–835.
- Holsinger JR. 1994. Pattern and process in the biogeography of subterranean amphipods. Hydrobiologia. 287(1):131–145.
- Lee C-W, Nakano T, Tomikawa K, Min G-S. 2018. The complete mitochondrial genome of Pseudocrangonyx daejeonensis (Crustacea: Amphipoda: Pseudocrangonyctidae). Mitochondrial DNA Part B. 3(2):823–824.
- Lee C-W, Tomikawa K, Nakano T, Min G-S. 2020. A new species of the genus Pseudocrangonyx (Crustacea: Amphipoda: Pseudocrangonyctidae) from Simbok Cave. Korea. Zootaxa. 4731(3):zootaxa.4731.3.2–334.
- Li JY, Liao YW, Li J, He LS. 2020. The complete mitochondrial genome of the deep-sea amphipod Eurythenes magellanicus (Crustacea: Amphipoda: Lysianassidae). Mitochondrial DNA Part B. 5(1):337–339.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Sidorov D, Holsinger JR. 2007. Procrangonyx stygoedincus, a new species of subterranean amphipod (Pseudocrangonyctidae) from the Far East of Russia, with remarks on biogeographic relationships. Crustac. 80(4):417–430.
- Song J-H, Kim S, Shin S, Min G-S. 2016. The complete mitochondrial genome of the mysid shrimp, Neomysis japonica (Crustacea, Malacostraca, Mysida). Mitochondrial DNA A DNA Mapp Seq Anal. 27(4):2781–2782.
- Tomikawa K, Nakano T. 2018. Two new subterranean species of Pseudocrangonyx Akatsuka & Komai, 1922 (Amphipoda: Crangonyctoidea: Pseudocrangonyctidae), with an insight into groundwater faunal relationships in western Japan. J Crustacean Biol. 38(4):460–474.
- Zhao S, Hou Z. 2017. A new subterranean species of Pseudocrangonyx from China with an identification key to all species of the genus (Crustacea, Amphipoda, Pseudocrangonyctidae). ZooKeys. 647:1–22.