Abstract
Lirianthe hodgsonii is a tree species of Magnoliaceae as least concern. In the present paper, the complete chloroplast genome (cpDNA) and basic annotated information of L. hodgsonii were reported and its phylogenetic relationship with other species in Magnoliaceae was analyzed. The size of its complete cpDNA is 159,693 bp, with a typical quadripartite structure comprising a pair of inverted repeat (IR) regions of 26,546 bp, a large single-copy (LSC) region of 87,848 bp and a small single-copy (SSC) region of 18,753 bp. The genome contains 131 unique genes, including 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The phylogenetic analysis showed that L. hodgsonii is affinal to Lirianthe bidoupensis and they form a monophyletic group with other seven Lirianthe species. This Lirianthe clade is sister to the Dugandiodendron and Talauma clade with high support. All genera mentioned in this analysis are monophyletic under the system of Magnoliaceae by Sima and Lu.
Lirianthe hodgsonii (J. D. Hooker & Thomson) Sima & S. G. Lu was initially described as a species of the genus Talauma Jussieu within the family Magnoliaceae for its circumscissile mature carpels by Hooker and Thomson (Citation1855). They were followed by some scholars (Law and Wu Citation1996; Xia et al. Citation2008) although it was ever transferred to the genus Magnolia Linnaeus by Keng (Citation1978). Based on further observations, Sima and Lu (Citation2009) renamed it Lirianthe hodgsonii (J. D. Hooker & Thomson) Sima & S. G. Lu. The species is native to SW China (S Xizang, SE and SW Yunnan), Bangladesh, Bhutan, NE India, N Myanmar, Nepal, and Thailand (Law and Wu Citation1996; Xia et al. Citation2008; Kundu Citation2009; Nooteboom and Chalermglin Citation2009; Sima and Lu Citation2009). It is a subdominant tree species in tropical moist forest (Barua et al. Citation2018) and used medicinally (Kijjoa Citation2002; Nascimento et al. Citation2004). In 2007, it was evaluated as least concern in the book, The Red List of Magnoliaceae (Cicuzza et al. Citation2007). However, there has been no report on chloroplast genome information of Lirianthe hodgsonii (J. D. Hooker & Thomson) Sima & S. G. Lu until now.
In the study, the complete sequence of chloroplast genome of Lirianthe hodgsonii (J. D. Hooker & Thomson) Sima & S. G. Lu was reported. The GenBank accession number is MT560391. The leaf sample of a tree of Lirianthe hodgsonii (J. D. Hooker & Thomson) Sima & S. G. Lu was collected from Motuo County, Xizang Province of China. The sheets of vouchered specimen, H. Jiang 7337, are stored at the herbarium, YAF. Genomic DNA was extracted from dry leaves of the specimen by using DNA Plantzol Reagent (Invitrogen, Carlsbad, CA). Total genome DNA of this species was sequenced by Illumina HiSeq Sequencing System (Illumina, San Diego, CA) and shotgun library was constructed. About 2.0 Gb pair-end (150 bp) raw sequence data were obtained and the low-quality sequences were filtered using CLC Genomics Workbench v8.0 (CLC Bio, Aarhus, Denmark) to get high-quality clean reads. NOVOPlasty software (Dierckxsens et al. Citation2017) was used to align and assemble cp genome with Pachylarnax sinica (Y. W. Law) N. H. Xia & C. Y. Wu (JX280400) served as the reference. The complete chloroplast genome of Lirianthe hodgsonii (J. D. Hooker & Thomson) Sima & S. G. Lu was automatically annotated using CpGAVAS (Liu et al. Citation2012) and then adjusted and confirmed with Geneious 9.1 (Kearse et al. Citation2012). Then, the annotated genomic sequence was submitted to GenBank.
The size of complete chloroplast genome of Lirianthe hodgsonii (J. D. Hooker & Thomson) Sima & S. G. Lu is 159,693 bp, with a typical quadripartite structure including a large single-copy (LSC) region of 87,848 bp and a small single-copy (SSC) region of 18,753 bp separated by a pair of inverted repeat (IR) regions of 26,546 bp each. The chloroplast genome contains 131 unique genes, including 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes.
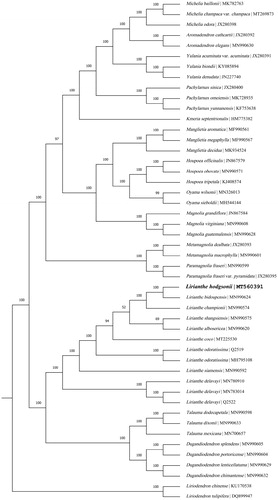
In order to determine the phylogenetic position of Lirianthe hodgsonii (J. D. Hooker & Thomson) Sima & S. G. Lu, 47 complete chloroplast genome sequences of the subfamily Magnolioideae from NCBI were aligned using MAFFT v. 7 (Sima and Lu Citation2012; Katoh and Standley Citation2013). Based on the system of Magnoliaceae by Sima and Lu (Citation2012), two species of the subfamily Liriodendroideae, Liriodendron chinense (Hemsley) Sargent (KU170538) and Liriodendron tulipifera Linnaeus (DQ899947) were served as the outgroup. The maximum-likelihood (ML) tree was reconstructed with RAxML (implemented in Geneious ver.10.1 http://www.geneious.com, Kearse et al. Citation2012) and bootstrap values were calculated from 1000 replicates (). The result of phylogenetic analysis revealed that Lirianthe hodgsonii (J. D. Hooker & Thomson) Sima & S. G. Lu is affinal to Lirianthe bidoupensis (Q. N. Vu) Sima & Hong Yu (MN990624) and formed a monophyletic group with the latter and other seven species of the genus Lirianthe Spach. This clade of the genus Lirianthe Spach is sister to the clade of the genus Dugandiodendron Lozano-Contreras and the genus Talauma Jussieu with high support. All genera mentioned in this analysis are monophyletic under the system of Magnoliaceae by Sima and Lu (Citation2012). The determination of the complete plastid genome sequences provided new molecular data to illuminate the genus Lirianthe Spach in Magnoliaceae evolution.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the result of this study are openly available in NCBI GenBank database (https://www.ncbi.nlm.nih.gov) and the accession number is MT560391, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Additional information
Funding
References
- Barua KN, Gogoi G, Hazarika P. 2018. Comparative study on structural composition and community association of Nambor Wildlife Sanctuary and its South-Westward extended Bornewria forest, Assam, India. Trop Plant Res. 5(2):233–242.
- Cicuzza D, Newton A, Oldfield S. 2007. The red list of Magnoliaceae. Cambridge: Fauna & Flora International.
- Dierckxsens N, Mardulyn P, Smits G. 2017. Novoplasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Hooker JD, Thomson T. 1855. Magnoliaceae. In: Flora Indica, 1. London: W. Pamplin; p. 72–79.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Keng H. 1978. The delimitation of the genus Magnolia (Magnoliaceae). Gard Bull Singapore. 31:127–131.
- Kijjoa A. 2002. Plant secondary metabolites with immunomodulatory activity. In: Rauter AP, editor. Natural products in the new millennium: prospects and industrial application. Vol. 47. Netherlands: Kluwer Academic Publishers; p. 299–309.
- Kundu SR. 2009. A synopsis on distribution and endemism of Magnoliaceae s.l. in Indian subcontinent. Thaiszia. 19(1):47–60.
- Law Y-W, Wu Y-F. 1996. Magnoliaceae. In: Flora Reipublicae Popularis Sinicae. Vol. 30. Beijing: Science Press; p. 82–198.
- Liu C, Shi LC, Zhu YJ, Chen HM, Zhang JH, Lin XH, Guan XJ. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715.
- Nascimento MSJ, Pedro M, Cerqueira F, Bastos M, Vieira LM, Kijjoa A, Pinto MM. 2004. Effect of natural 2,5-diaryl-3,4-dimethyltetrahydrofuran lignans on complement activation, lymphocyte proliferation, and growth of tumor cell lines. Pharm Biol. 42(6):449–453.
- Nooteboom HP, Chalermglin P. 2009. The Magnoliaceae of Thailand. Thai Bull (Bot). 37:111–138.
- Sima YK, Lu SG. 2009. Magnoliaceae. In: Shui YM, Sima YK, Wen J, Chen WH, editors. Vouchered flora of Southeast Yunnan. Vol. 1. Kunming: Yunnan Science & Technology Press; p. 16–67.
- Sima YK, Lu SG. 2012. A new system for the family Magnoliaceae. In: Xia NH, Zeng QW, Xu FX, Wu QG, editors. Proceedings of the Second International Symposium on the Family Magnoliaceae. Wuhan: Huazhong University Science & Technology Press; p. 55–71.
- Xia NH, Liu Y, Law YW, Nooteboom HP. 2008. Magnoliaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 7. Beijing/St. Louis: Science Press/Missouri Botanical Garden Press; p. 48–91.