Abstract
This study reports the complete mitochondrial genome of Squalidus multimaculatus. The length of the mitochondrial genome of S. multimaculatus is 16,597 bp, including 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes, and one control region. The overall base composition of A, G, T, and C is 28%, 18.8%, 25%, and 28.2%, respectively. A phylogenetic analysis by the neighbour-joining method showed a close relationship between S. multimaculatus and S. japonicus coreanus. We believe these results will provide essential data for phylogeographic studies of the genus Squalidus.
Squalidus multimaculatus, S. chankaensis tsuchigae, S. japonicus coreanus, and S. gracilis majimae of the Squalidus (Cypriniformes: Cyprinidae) genus are endemic freshwater species that are distributed in South Korea. S. multimaculatus has a restricted habitat in the rivers flowing to the East coast and is threatened with extinction (Lee et al. Citation2017).
In this study, we described the characterization of the complete mitochondrial genome of S. multimaculatus and analyzed its phylogenetic relationships within the genus Squalidus. Our study will aid in the sustainable management of this species and facilitate future research on taxonomic resolution, population genetic structure, and phylogenetic relationships.
Squalidus multimaculatus was collected from Gokgangcheon River, Pohang-si (N36.074631°, E129.165850°) in 2017. The voucher specimen has been archived at the Molecular Phylogenetics Laboratory in Nakdonggang National Institute of Biological Resources, Sangju-si, Korea under the accession number NNIBR-MPL2017GSM0013. The mitogenome sequence was extracted from the next-generation sequencing data (unpublished data) using the Illumina HiSeq 4000 platform by GnC Bio (Daejeon, Korea). In total, 158 Gb raw reads were obtained and assembled using the Platanus assembler (ver.1.2.4., Kajitani et al. Citation2014). In total, 77,028 reads were used to reconstruct the mitogenome using Deconseq (ver.0.4.3., Schmieder and Edwards Citation2011) and it was annotated using gsMapper (ver. 2.8.; Roche) using MitoFish database as the reference (Sato et al. Citation2018).
The complete mitogenome of S. multimaculatus is 16 597 bp (GenBank accession number: MK840865) in length, consisting of 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and one control region (D-loop). The control region was 924 bp in length and located between the tRNAPro and tRNAPhe. The overall base composition was 28% of A, 18.8% of G, 25% of T, 28.2% of C, and had a slight AT bias of 53%. All the PCGs used the start codon ATG except COI, which used GTG; this is in accordance with the mitochondrial genome analysis of 250 fishes performed by Satoh et al. Five genes (COI, ATPase8, ND4L, ND5, and ND6) used TAA as a stop codon, ND1 used TAG as a stop codon, whereas the remaining seven genes used incomplete stop codons (T–– or TA–). It is speculated that having 3′ end PCGs followed by the tRNA gene encoded on the same strand may permit transcription to terminate without complete stop codons (Satoh et al. Citation2016). The 12S and 16S rRNAs were 959 bp and 1685 bp, respectively. The tRNA sequence length ranged from 69 to 76 bp; all the tRNA genes, which were predicted using MITOS (Bernt et al. Citation2013), had a three-leaf clover structure, except tRNA-SerUCG.
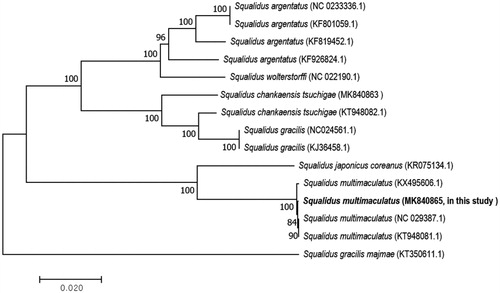
To examine the phylogenetic position of S. multimaculatus, we downloaded the mitochondrial genome sequences of five species belonging to the genus Squalidus from GenBank (Tang et al. Citation2011; Li et al. Citation2016; Liu et al. Citation2016; Zhou et al. Citation2016; Park et al. Citation2016, Citation2020; Yi et al. Citation2017). The phylogenetic tree was constructed with the sequences of the 13 PCGs using the neighbor joining method in the program MEGA7. A close relationship was observed between S. japonicas coreanus and S. multimaculatus (KX495606.2, and in this study; ). Squalidus multimaculatus has a restricted habitat, hence, more genetic and phylogeographic studies are needed to preserve this species. Our study provides useful information that may contribute toward future phylogeographic studies in the Squalidus species.
Figure 1. A phylogenetic tree construction of five species based on 13 protein-coding gene sequences of complete mitochondrial genome sequences accessed from GenBank using the neighbor-joining method. The tree was computed using Kimura’s 2-parameter distance model with bootstrap value: 10,000. The scale bar indicates 0.02 substitutions per nucleotide position.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available in GenBank, NCBI at https://www.ncbi.nlm.nih.gov under the accession number: MK840865.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, Okuno M, Yabana M, Harada M, Nagayasu E, Maruyama H, et al. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24(8):1384–1395.
- Lee YJ, Bae H-G, Jeon H-B, Kim D-Y, Suk HY. 2017. Human-mediated processes affecting distribution and genetic structure of Squalidus multimaculatus, a freshwater cyprinid with small spatial range. Animal Cells and Systems. 21(5):349–357.
- Li J, Huang L, Wang J, Chen Y-J, Du H-D, Huang T-J. 2016. The complete mitochondrial genome of Squalidus argentatus (Cypriniformes, Cyprinidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):38–39.
- Liu G-D, Chen IS, Zhu J-Q, Shen C-N. 2016. The complete mitochondrial genome of small sliver gugeon Squalidus gracilis (Teleostei, Cyprinidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):603–604.
- Park SY, Kim SK, Sagong J, Ryu S-H, Yu J-N. 2020. Complete mitochondrial genome sequence of Squalidus chankanensis tsuchigae (Cypriniformes: Cyprinidae). Mitochondrial DNA Part B. 5(1):412–413.
- Park CE, Park G-S, Kwak Y, Kim M-C, Kim K-H, Park HC, Shin J-H. 2016. The complete mitochondrial genome sequence of Squalidus japonicus coreanus (Teleostei, Cypriniformes, Cyprinidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3723–3724.
- Sato Y, Miya M, Fukunaga T, Sado T, Iwasaki W. 2018. MitoFish and MiFish pipeline: a mitochondrial genome database of fish with an analysis pipeline for environmental DNA metabarcoding. Mol Biol Evol. 35(6):1553–1555.
- Satoh TP, Miya M, Mabuchi K, Nishida M. 2016. Structure and variation of the mitochondrial genome of fishes. BMC Genomics. 17:719.
- Schmieder R, Edwards R. 2011. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS One. 6(3):e17288.
- Tang KL, Agnew MK, Chen WJ, Vincent Hirt M, Raley ME, Sado T, Schneider LM, Yang L, Bart HL, He S, et al. 2011. Phylogeny of the gudgeons (Teleostei: Cyprinidae: Gobioninae). Mol Phylogenet Evol. 61(1):103–124.
- Yi S, Lee S-H, Park T, Jang M-H. 2017. The complete mitochondrial genome sequence of Squalidus multimaculatus (Cypriniformes: Cyprinidae). Mitochondrial DNA B. 2(2):411–412.
- Zhou Q, Hu Y-X, Ye Q, Xue Y, Chen D-Q, Li Y. 2016. Mitochondrial genome of silver gudgeon, Squalidus argentatus (Teleostei, Cypriniformes). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):151–152.
