Abstract
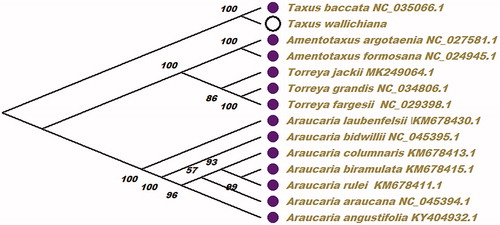
Taxus wallichiana is a member of the family Taxaceae, which is a unique and endangered species in China and is widely used for ornamental, material and medicinal purposes. The complete chloroplast genome of T. wallichiana was found to possess a total size of 128,168 bp. The GC content of T. wallichiana chloroplast genome sequence is 37.3%, the overall nucleotide composition of chloroplast genome sequence is: A of 30.7%, T of 32.0%, C of 19.0% and G of 18.3%. The total of 116 genes were successfully annotated, which contained 83 protein-coding genes, 29 transfer RNA genes, and 4 ribosomal RNA genes. The ML phylogenetic analysis result showed that T. wallichiana was closely related to Taxus baccata in the phylogenetic relationship using the neighbour-joining (NJ) method in this study.
Taxus wallichiana is the IUCN Red List species in the world, which is also one of the most important tree and medicinal plant species in China. Taxus wallichiana is one of the slow growing family Taxaceae species that is found to be the major source of Taxol used as the anti-cancer agent (Sudina and Dhurva Citation2018). Taxol is originally isolated from the bark of T. wallichiana, which is one of the most effective antitumor drugs for the treatment of several cancers, such as breast, lung and ovarian cancers (Croteau et al. Citation2006). Currently, very less is known about the biology and genomics information of T. wallichiana. In this paper, the complete chloroplast genome of T. wallichiana was characterized and generated that can be used for genome information resource collection and further drug development research for this species.
Taxus wallichiana fresh samples were collected from the Kunming Medical University, which were located at 102.83E, 24.85 N (Kunming, Yunnan, China) and stored at the −80 °C refrigerator future use. The Taxus wallichiana genomic DNA was extracted from the fresh leaves using the modified CTAB method, which was stored in Kunming Medical University (No.KMMU-001) and sequenced. Furthermore, the reads quality were controlled and removed using the FastQC (Andrews Citation2015). The chloroplast genome sequence of T. wallichiana was employed to obtain and assemble using the MitoZ (Meng et al. Citation2019). The chloroplast genome annotation was conducted using Geneious (Kearse et al. Citation2012). All the coding and other genes were predicted using the CPGAVAS (Liu et al. Citation2012) for this chloroplast genome. At last, the circular chloroplast genome map of T. wallichiana was generated using OrganellarGenomeDRAW online (Greiner et al. Citation2019).
The complete chloroplast genome of T. wallichiana (NCBI No.NK9584421) was found to possess a total size of 128,168 bp. The GC content of T. wallichiana chloroplast genome sequence is 37.3%, the overall nucleotide composition of chloroplast genome sequence is: A of 30.7%, T of 32.0%, C of 19.0% and G of 18.3%. The total of 116 genes were successfully annotated, which contained 83 protein-coding genes, 29 transfer RNA genes, and 4 ribosomal RNA genes.
To ascertain the position of T. wallichiana within other plant species, the maximum-likelihood (ML) phylogenetic tree was reconstructed using 13 species complete chloroplast genome sequence from NCBI (). In this paper, the ML phylogenetic tree was reconstructed using PhyML (Guindon et al. Citation2010) with 200 bootstrap replicates and with GTR + G + I as the best-fit nucleotide substitution model. At last, the ML phylogenetic tree was edited using MEGA X (Kumar et al. Citation2018). The ML phylogenetic analysis result showed that T. wallichiana was closely related to Taxus baccata in the phylogenetic relationship in this study. The result can be used for the use of active ingredients of traditional Chinese medicine and drug development research in future.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request. The data that support the findings of this study are openly available in Taxus wallichiana at NCBI and http://doi.org/10.1080/23802359.2020.1799725, reference number [reference number NK9584421].
Additional information
Funding
References
- Andrews S. 2015. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Croteau R, Ketchum REB, Long RM, Kaspera R, Wildung MR. 2006. Taxol biosynthesis and molecular genetics. Phytochem Rev. 5(1):75–97.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715–2164.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Sudina B, Dhurva PG. 2018. Taxus wallichiana (Zucc.), an endangered anti-cancerous plant: a review. Int J Res. 5(21):10–21.