Abstract
Some populations of the East Asian minnow Pungtungia herzi Herzenstein, 1892, which are naturally distributed in western Japan, have declined and are considered endangered. However, P. herzi has become a domestic invasive species in eastern Japan. Thus, knowledge of genetic features and phylogenetic relationships of P. herzi is important for conservation of this species and understanding its impact on ecosystems. We analyzed the complete mitochondrial genome using next generation sequencing of the East Asian minnow P. herzi from Yodo River, Osaka Prefecture, Japan. The mitochondrial genome of P. herzi consists of a circular molecule of 16,599 bp that includes 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and one control region. The heavy (H)-strand was predicted to have 12 PCGs, 14 tRNA, and two rRNA genes, while the light (L)-strand was predicted to contain one PCGs and eight tRNA genes. The average AT content was 57.68%. The genes ATP8 and ATP6, ATP6 and COIII, ND4L and ND4, and ND5 and ND6 shared seven, one, seven, and four nucleotides, respectively. The initiation codons ATG and GTG were found in 12 and one genes, respectively. The termination codons TAA, TAG, incomplete TA–, and single T–– were observed in nine, one, one, and two genes, respectively. All the tRNA genes possessed a cloverleaf secondary structure. The phylogenetic relationships inferred using 13 PCGs (based on the maximum likelihood) were consistent with previous studies that predicted interrelationships of Cypriniformes.
Pungtungia herzi Herzenstein, 1892 is known for parasitizing other fish species (e.g. Baba and Karino Citation1998; Baba Citation2010) and is distributed across the Korean Peninsula and Japan. In Japan, P. herzi is naturally distributed towards west from Fukui, Gifu, and Mie Prefectures (Yashima et al. Citation2011; Ohnaka and Mukai Citation2019). Populations of P. herzi have declined in some of these areas (e.g. Ito and Morimoto Citation2003; Osaka Prefecture Citation2014). However, P. herzi is a domestic invasive species, mainly in the Kanto region, and is a concern because of its influence on ecosystems (e.g. Yashima et al. Citation2011). Hence, understanding the genetic features and phylogenetic relationships between P. herzi populations and closely related species is important. Here, we present the complete mitochondrial genome of P. herzi from Osaka Prefecture, Japan, where it is being designated as an endangered species (Osaka Prefecture Citation2014).
Pungtungia herzi were captured alive from Yodo River, Osaka Prefecture, Japan, and stored in the Lake Biwa Museum (accession number: LBM-1210058083). DNA samples were immediately extracted using NucleoSpin Tissue (MACHEREY-NAGEL) and stored in a freezer at −20 °C for mitochondrial DNA analysis. Genomic DNA isolated from one fish was sequenced using the Illumina MiSeq platform (Illumina, San Diego, CA, USA). The complete mitochondrial genome of P. herzi (AB239598; Saitoh et al. Citation2006) was used as a reference sequence. The resultant reads were assembled and annotated using the MITOS web server (Bernt et al. Citation2013) and Geneious R9 (Biomatters) software. Thirteen protein-coding genes (PCGs) and two rRNA genes sequences were aligned using MEGAX (Kumar et al. Citation2018). The phylogenetic analysis was performed with the maximum likelihood (ML) criterion using TREEFINDER version of March 2011 (Jobb Citation2011).
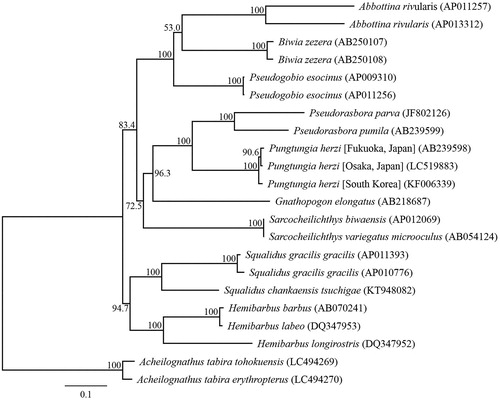
We succeeded in sequencing the entire mitochondrial genome of P. herzi from Osaka Prefecture (DDBJ accession number LC519883). The genome resembles the genomic organization common in P. herzi and consisted of a closed loop that was 16,599 bp-long, which included 13 PCGs, 22 tRNA genes, two rRNA genes, and one control region. The average AT content was 57.68%. The heavy strand was predicted to have 12 PCGs, 14 tRNA, and two rRNA genes, while the light strand was predicted to contain one PCG and eight tRNA genes. Among the PCGs, ATP8 and ATP6, ATP6 and COIII, ND4L and ND4, and ND5 and ND6 shared seven, one, seven, and four nucleotides, respectively. Twelve PCGs of the P. herzi mitochondrial genome started with ATG, the COI gene started with GTG, and the termination codons TAA and TAG were observed in nine PCGs and the ND4 gene, respectively. The incomplete stop codons TA (COIII) and T (COII and Cytb) were identified. All tRNA genes possessed a cloverleaf secondary structure. A phylogenetic analysis was conducted using the sequence information of 13 mitochondrial PCGs from 15 Japanese Gobioninae taxa (). Two Japanese P. herzi formed a sister clade. The phylogenetic relationships of Japanese Gobioninae taxa were consistent with a previous study, which predicted interrelationships of the Cypriniformes (Saitoh et al. Citation2006; Tang et al. Citation2011).
Figure 1. Phylogenetic relationships (maximum likelihood) of the Japanese Gobioninae taxa based on the nucleotide sequences of the 13 protein-coding genes of the mitochondrial genome. Sequences from Acheilognathus tabira tohokuensis (LC494269) and A. tabira erythropterus (LC494270) were used as an outgroup (Nagata and Kitamura Citation2019). These sequences were separated by codon positions, and for each partition, the optimal models of sequence evolution were used in the maximum likelihood method using TREEFINDER, based on the corrected Akaike information criterion. The numbers at the nodes indicate the bootstrap support inferred from 1000 bootstrap replicates. Alphanumeric terms indicate the DNA Database of Japan accession numbers.
Acknowledgements,
We are grateful to staff and Science Research Club members of Osaka High School for kindly support for helping our research.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available on GenBank using the accession numbers LC519883 (https://www.ncbi.nlm.nih.gov/nuccore/LC519883).
Additional information
Funding
References
- Baba R, Karino K. 1998. Countertactics of the Japanese aucha Perchsiniperca kawamebari against brood parasitism by the Japanese minnow Pungtungia herzi. J Ethol. 16(2):67–72.
- Baba R. 2010. Timing of spawning and host-nest choice for brood parasitism by the Japanese minnow, Pungtungia herzi, on the Japanese aucha perch, Siniperca kawamebari. Ethology. 98(1):50–59.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Ito S, Morimoto Y. 2003. Garden ponds as wildlife habitats for fish from Lake Biwa into Kyoto city. J Japanese Inst Landscape Architecture. 66(5):621–626. (In Japanese).
- Jobb G. 2011. TREEFINDER version of March 2011. Munich. http://www.treefnder.de.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Nagata N, Kitamura JI. 2019. The complete mitochondrial genomes of two endangered bitterling Acheilognathus tabira tohokuensis and A. tabira erythropterus (Cyprinidae, Acheilognathinae). Mitochondrial DNA Part B. 4(2):2865–2866.
- Ohnaka T, Mukai T. 2019. First record of non-indigenous freshwater fish, Pungtungia herzi (Cypriformes: Cyprinidae) collected from Inuyama City, Aichi Prefecture. Sci Rep Toyohashi Museum Nat Hist. 29:33–35. (In Japanese).
- Osaka Prefecture. 2014. Red list of Osaka Prefecture 2014: Osaka Prefectural Government. http://www.pref.osaka.lg.jp/attach/21490/00148206/zentai.pdf. (In Japanese).
- Saitoh K, Sado T, Mayden RL, Hanzawa N, Nakamura K, Nishida M, Miya M. 2006. Mitogenomic evolution and interrelationships of the Cypriniformes (Actinopterygii: Ostariophysi): the first evidence toward resolution of higher-level relationships of the world's largest freshwater fish clade based on 59 whole mitogenome sequences. J Mol Evol. 63(6):826–841.
- Tang KL, Agnew MK, Chen WJ, Hirt MV, Raley ME, Sado T, Schneider LM, Yang L, Bart HL, He S, et al. 2011. Phylogeny of the gudgeons (Teleostei: Cyprinidae: Gobioninae). Mol Phylogenet Evol. 61(1):103–124.
- Yashima N, Tamino T, Kitano T. 2011. Notes of Pungtungia herzi and Noemacheilus barbatulus toni as domestic alien species collected from Kaname River. Nat Hist Rep Kanagawa. 32:109–113.
