Abstract
Dipterocarpus tempehes is an endangered tropical timber tree species endemic to Borneo. In this study, the plastome of D. tempehes was assembled. The whole chloroplast genome was 145,323 bp in length, with a large single-copy (LSC) region of 80,375 bp and small single-copy (SSC) region of 20,394 bp, separated by two inverted repeats (IRs) regions of 22,277 bp. It contained 145 genes, including 102 coding genes, 35 tRNA genes, and 8 rRNA genes. A phylogenomic analysis confirmed that Malvaceae s.l., Thymelaeaceae, and Dipterocarpaceae formed its respective monophyletic clade, with D. tempehes forming a sister clade with species from the Shoraeae tribe within the family.
Dipterocarpus tempehes Slooten is an emergent tree species from the lowland tropical rainforests of Borneo. It is an endangered species on the IUCN Red List (Bodos et al. Citation2019) with high economic value as an important timber species and ecological value as a key structural component of the forest. Here, we report on a newly sequenced plastome of D. tempehes and its phylogenomic relationships with 15 other species from closely-related families. The genomic data provided can be further used for population genomic, phylogenomic reconstruction, and genetic engineering studies of the genus Dipterocarpus and the family Dipterocarpaceae.
Total genomic DNA was extracted from fresh leaves of a D. tempehes tree using the Plant Genomic DNA Kit (Tiangen Biotech Co., Ltd). The individual was collected from northern Kalimantan, Indonesia (Latitude 3°28′51,5′′, Longitude 116°35′25,3′′), planted in Bali Botanical Garden, Indonesia, and the voucher sample was deposited in Herbarium Hortus Botanicus Baliense, Bali Botanical Garden (RS05) and the Ecological Genomic Team tissue bank (EGT), Guangxi University, Nanning, China. Library construction and sequencing were performed by Novogene (Nanjing, China). The draft genome was reconstructed with NOVOPlasty version 2.7.0 using default settings. The raw reads and resulting circular contigs were imported in Geneious R11 version 11.0.4 (Dierckxsens et al. Citation2016) for further assembly and plastome reconstruction. The plastome of another D. tempehes individual (BGT4271, unpublished data; vouchers deposited in the Biodiversity Genomics Team herbarium (BGT), Nanning, China) was used as a mapping reference in Geneious R11 version 11.0.4 (https://www.geneious.com, Kearse et al. Citation2012) and annotations were determined using GeSeq (Tillich et al. Citation2017).
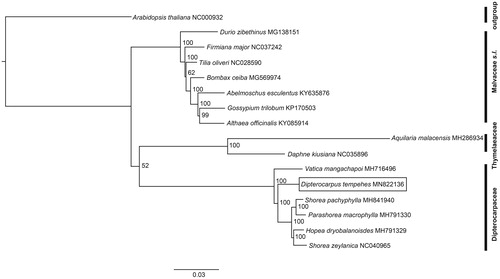
A phylogenetic tree was constructed using the plastomes of D. tempehes and 14 other species from Malvales (seven species from Malvaceae s.l., two species from Thymelaeaceae, and five species from Dipterocarpaceae), with Arabidopsis thaliana as an outgroup, all downloaded from GenBank. We used ModelTest-NG version 0.1.5 (Darriba et al. Citation2020) to find the best fitting model for our dataset (GTR + I + G4). A maximum likelihood (ML) tree was built using RAxML-NG version 0.8.1 (Kozlov et al. Citation2019), using standard bootstrap analyses with 1000 replicates to evaluate node supports (Kozlov et al. Citation2019). The ML tree was edited in FigTree version 1.4.3 (Rambaut Citation2017).
The plastome of D. tempehes (GenBank accession MN822136) was 145,323 bp in length, consisting of a large single-copy (LSC) with 80,375 bp, a small single-copy (SSC) region of 20,394 bp, separated by two inverted repeats (IRs) regions of IR 22,277 bp. The GC content in the chloroplast genome was 43.3% in the IRa and 36.8% in the IRb, which was greater than that of the LSC (35.3%) and SSC (31.4%) regions.
The phylogenomic analysis showed a clearly resolved relationship among analyzed families in Malvales: Malvaceae s.l., Thymelaeaceae, and Dipterocarpaceae formed a monophyletic clade, respectively. Dipterocarpaceae and Thymelaeaceae formed a sister clade, with Malvaceae s.l being more distantly related (). This is in agreement with previously published phylogenomic analysis using chloroplast genomes of Malvales (Yan et al. Citation2019). Within Dipterocarpaceae, D. tempehes formed a sister clade with species from the Shoraeae tribe (Hopea, Parashorea, and Shorea), with V. mangachapoi in a basal position. This lent further support to a recently available phylogeny of Dipterocarpaceae using complete chloroplast genomes (Cvetković et al. Citation2017, Citation2019). The plastome of D. tempehes is a useful resource for conservation genetic studies of this endangered species, especially in resolving the phylogenomic relationships and evolutionary history of the mega-diverse Dipterocarpaceae.
Acknowledgments
We thank Assoc. Prof. Joeri S. Strijk from Guangxi University for his guidance in data analysis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in Genbank of NCBI at http://www.ncbi.nlm.nih.gov.
Additional information
Funding
References
- Bodos V, Kusumadewi Y, Robiansyah I, Randi A, Tsen S, Pereira J, Maycock CR. 2019. Dipterocarpus tempehes. The IUCN red list of threatened species. T33378A125628354. [accessed 2020 Jun 30]. https://dx.doi.org/10.2305/IUCN.UK.2019-3.RLTS.T33378A125628354.en
- Cvetković T, Hinsinger DD, Strijk JS. 2017. Exploring the first complete chloroplast sequence of a major tropical timber tree in the Meranti family: Vatica odorata (Dipterocarpaceae). Mitochondrial DNA Part B. 2(1):52–53.
- Cvetković T, Hinsinger DD, Strijk JS. 2019. Exploring evolution and diversity of Chinese Dipterocarpaceae using next-generation sequencing. Sci Rep. 9(1):11639.
- Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T. 2020. Model test-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol Evol. 37(1):291–294.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVO Plasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21):4453–4453.
- Rambaut A. 2017. Figtree-version 1.4.3, a graphical viewer of phylogenetic trees. Computer program distributed by the author, website: http://tree. bio. ed. ac. uk/software/figtree
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Yan F, Tao X, Wang QL, Juan ZY, Zhang CM, Yu HL. 2019. The complete chloroplast genome sequence of the medicinal shrub Daphne giraldii Nitsche. (Thymelaeaceae). Mitochondrial DNA Part B. 4(2):2685–2686.