Abstract
Pinus Yunnanensis var. pygmaea (Fam: pinaceae; Gen: Pinus), is a mutant of Pinus yunnanensis. Franch. To contribute to its conservation, the complete chloroplast (cp) genome of P. yunnanensis var. Pygmaea was sequenced and assembled by high-throughput sequencing technology. The results show that P. yunnanensis var. pygmaea cp genome contained 101 genes, including 64 protein-coding genes, 33 tRNA genes, and four rRNA genes. The overall GC content of the cp genome is 38.50%. A phylogenetic tree reconstructed by 16 cp genomes reveals that P. yunnanensis var. pygmaea is most related with Pinus taiwanensis.
Pinus Yunnanensis var. pygmaea (Fam: pinaceae; Gen: Pinus), is a mutant of Pinus yunnanensis. Franch., distributed in Muli and Zhaojue in southwestern Sichuan Province and the mountains of northwestern and central Yunnan Province, China. The P. pygmaea (P. yunnanensis var. pygmaea) is inconspicuous, and the basal part has many trunks, which are clustered, cones are clustered, which are persistent on the tree after ripening; the fruiting phenomenon is generally early (Zheng-li et al. Citation1994; Yu-Lan et al. Citation2012).
At present, cp genome, mitochondrial genome, and nucleus genome are three major genomes used for phylogenetic studies in plant, among which cp genome is widely used in the study of kinship between different species and species evolution because of its conserved number and sequence of genes (Njuguna et al. Citation2013).
The P. pygmaea seeds were collected from Kunming City, Yunnan Province, China (E:103°13′; N:24°52′), and kept at the Institute of Forest Resources & Environment Center of Guizhou Province, the deposit number is NO. PY-001-1. Collected annual new needles for Illumina NovaSeq sequencing to obtain the cp genome. The P. pygmaea cp genome sequence was obtained by SPAdes (Bankevich et al. Citation2012) and A5-miseq (Coil et al. Citation2015) splicing fragments. The annotated cp genome sequence has been deposited into the Genbank (accession: MT712078).
P. pygmaea cp genome was not distinctly tetragonal with 119,570 bp in length. The content of GC is 38.50%, the annotated complete cp genome contains 101 genes, including 64 protein-coding gene, four ribosomal RNA, and 33 transfer RNA.
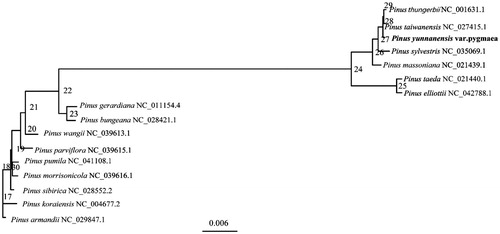
The phylogenetic relationships of 16 different species of the P. pygmaea were investigated using the evolutionary tree construction method using IQ-tree software (Nguyen et al. Citation2015) with maximum-likelihood (ML) method. A phylogenetic tree reconstructed by 16 cp genomes reveals that P. pygmaea is most related with Pinus taiwanensis (). The results of the phylogenetic relationships are consistent with the results of the morphological classification, laying the foundation for the study of the evolution of pinaceae species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT712078.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31(4):587–589.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Njuguna W, Liston A, Cronn R, Ashman TL, Bassil N. 2013. Insights into phylogeny, sex function and age of Fragaria based on whole chloroplast genome sequencing. Mol Phylogenet Evol. 66(1):17–29.
- Yu-Lan XU, Nian-Hui C, Xiang-Yang K, Gen-Qian LI, Cheng-Zhong HE, An-An D. 2012. Advances in the study on the genetic relationships of Pinus yunnanensis and its related species. J Northwest Forest Univ. 27(1):98–102.
- Zheng-Li L, Yong-Jun F, Ke-Ming C. 1994. Comparative anatomical observations of wood structures of Pinus yunnanensis and P. yunnanensis var. pygmaea. J Integr Plant Biol. 36(7):502–505.