Abstract
Koelreuteria bipinnata is an important ornamental tree with attractive flowers and fruits. In this study, we used next-generation sequencing technology to obtain the complete chloroplast genome of K. bipinnata. The entire genome was determined to be 163,863 bp in size, harboring a typical quadripartite structure with a large single copy (LSC) region of 90,240 bp, a small single copy (SSC) region of 18,883 bp, and a pair of 27,370 bp inverted repeat (IR) regions. The genome was predicted to contain 132 genes, including 84 protein-coding genes, 40 tRNA genes, and 8 rRNA genes. The overall GC content of K. bipinnata chloroplast genome was 37.29%. Phylogenetic analysis based on complete chloroplast genome sequences indicated that K. bipinnata was closely related to K. paniculate. This study would be useful for future population genetics studies and phylogenetic analysis of K. bipinnata.
Koelreuteria bipinnata, also named K. bipinnata Franch. var. integrifoliola (Merr.) T. Chen, is an extremely valuable tree species in Sapindaceae family (Feng, Li, et al. Citation2009). Traditionally, its dry flowers were used as herbal medicine for curing eye disease, and woods were utilized for furniture production in China. Nowadays, it has become a popular tree for urban landscaping as its attractive flowers and fruits (Lyu et al. Citation2017). Previously, the studies regarding K. bipinnata were mainly focus on its tissue culture (Feng, Li, et al. Citation2009; Feng, Zhang, et al. Citation2009; Cao et al. Citation2018), few data are currently available about its genomic information. In plants, the chloroplast genome is helpful for population genetics study, species-level identification, and phylogenetic analysis (Liu et al. Citation2020; Mo et al. Citation2020). Here, we de novo assembled the complete chloroplast genome of K. bipinnata, and analyzed its relationship with closely related species.
Leave samples of K. bipinnata was obtained from the experimental farm of Jiangsu Academy of Forestry (Nanjing, China 118°45′57.30″E, 31°51′27.94″N) and the voucher specimen was stored at Herbarium of Jiangsu Academy of Forestry (JAF; voucher: Lyu20200512-1). Total genomic DNA was extracted using Tiangen Plant Genomic DNA Kit (Tiangen Biotech Co., Beijing, China). The obtained DNA was fragmented to construct a paired-end library with an insert-size of 300 bp, and then the library was sequenced on the Illumina NovaSeq system (Illumina, San Diego, CA, USA). Following sequencing, 5.59 Gb of raw data were generated. Raw reads were processed by the Trimmomatic v0.32. De novo assembly and annotation of K. bipinnata chloroplast genome was performed by NOVOPlasty and DOGMA, respectively. The annotated cp genome was deposited in GenBank (Accession number: MT675915).
The whole chloroplast genome of K. bipinnata was 163,863 bp in size, and presented a typical quadripartite structure with a large single copy (LSC) region of 90,240 bp, a small single copy (SSC) region of 18,883 bp, and two separated inverted repeats (IRa and IRb) of 27,370 bp each. A total of 132 genes were predicted to include in the chloroplast genome: 84 protein-coding genes, 40 tRNA genes, and 8 rRNA genes. Of these genes, 7 protein-coding genes, 8 tRNA genes, and 4 rRNA genes have two copies, one in each of the IR regions. The overall GC content of the K. bipinnata chloroplast genome was 37.29%.
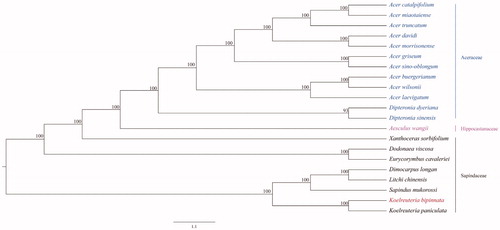
Phylogenetic tree was constructed using the chloroplast genome sequences of 21 representative Sapindales species: 12 from Aceraceae, 1 from Hippocastanaceae, and 8 from Sapindaceae. Multiple sequence alignment was completed with MAFFT (Katoh and Standley Citation2013) and then phylogenetic analysis was constructed using maximum-likelihood method implemented in IQ-tree software (Nguyen et al. Citation2015). The results showed that the clades in the phylogenetic tree have high bootstrap value, and species belonging to the same family were grouped together. Additionally, a close relationship was founded between K. bipinnata and K. paniculate. The complete chloroplast genome of K. bipinnata would be useful for its phylogenetic position determination as well as future population genomic studies ().
Figure 1. Phylogenetic tree constructed using ML method based on the complete chloroplast genome sequences of K. bipinnata and other 20 species. The bootstrap support values were shown at the branches. The accession numbers are the following: Acer amplum, NC_041080; Acer buergerianum, KF753631; Acer davidii, KU977442; Acer griseum, NC_034346; Acer laevigatum, NC_042443; Acer miaotaiense, KX098452; Acer morrisonense, KT970611; Acer sino-oblongum, NC_040106; Acer truncatum, NC_037211; Acer wilsonii, NC_040988; Aesculus wangii, NC_035955; Dimocarpus longan, NC_037447; Dipteronia dyeriana, KT985457; Dipteronia sinensis, KT878501; Koelreuteria paniculata, NC_037176.1; Sapindus mukorossi, KM454982; Xanthoceras sorbifolium, NC_037448; Dodonaea viscosa, NC_036099.1; Eurycorymbus cavaleriei, NC_037443; Litchi chinensis, NC_035238.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at http://www.ncbi.nlm.nih.gov/, reference number MT675915.
Additional information
Funding
References
- Cao L, Liu J, Lin Q, De Craene LPR. 2018. The floral organogenesis of Koelreuteria bipinnata and its variety K. bipinnata var. integrifolia (Sapindaceae): evidence of floral constraints on the evolution of monosymmetry. Plant Syst Evol. 304(8):923–935.
- Feng D, Li W, Zhang J, Li P, Li M, Zhao S, Shi B. 2009. Observation on somatic embryogenesis and histology in Koelreuteria bipinnata var. integrifoliola. Plant Physiol Commun. 45(9):855–858.
- Feng DL, Zhang J, Liu X, Peng WX, Wu TY. 2009. In vitro culture of immature embryos from Koelreuteria bipinnata var. integrifoliola. For Stud China. 11(3):179–184.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Liu J, Chen T, Zhang Y, Li Y, Gong J, Yi Y. 2020. The complete chloroplast genome of Rhododendron delavayi (Ericaceae). Mitochondr DNA Part B. 5(1):37–38.
- Lyu Y, Dong X, Huang L, Huang L. 2017. De novo assembly of Koelreuteria transcriptome and analysis of major gene related to leaf etiolation. S Afr J Bot. 113:355–361.
- Mo Z, Lou W, Chen Y, Jia X, Zhai M, Guo Z, Xuan J. 2020. The chloroplast genome of Carya illinoinensis: genome structure, adaptive evolution, and phylogenetic analysis. Forests. 11(2):207.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
