Abstract
Xyrias revulsus is one of the species in the China-Vietnam Collective Fishery Zone, which has a few relevant studies. In this study, the mitochondrial genome of X. revulsus was determined for the first time using next-generation sequencing; the overall base components of mitogenome consisting of 17,784 bp was 32.45% for A, 25.76% for T, 15.72% for G, 26.08% for C, and its GC content was 41.8%. The mitochondrial circular genome was composed of 13 protein-coding genes, 22 transfer RNAs, 2D-loop, and 2 ribosomal RNAs. Polygenetic analysis showed that the X. revulsus was very close to Ophisurus macrorhynchos. It can provide data reference for the analysis of genetic evolution of this species.
Xyrias revulsus is a member of Guilliformes, Ophichthidae, Xyrias, mainly distributed in the indo-pacific seas (Chen Citation2006). It is a rare species that inhabits waters from a few meters to 300 m depth (McCosker Citation1998). Little is known about it, which was first discovered in 1901 (Jordan and Snyder Citation1901). So far it is only reported in the east of Evans Shoal, the north of Bathhurst Island, the Izu Peninsula, Diaoyu Islands and the east China Sea (Huang and Zhan Citation1981; Tan Citation1983; Anonymous Citation1996; Mccosker et al. Citation2009). Little research has been done on the genome, population genetics and evolutionary relationships. Mitochondrial genomes has the advantages of stable structure and compact gene arrangement, which has been widely used in the study of Metazoan population genetics, biogeography and system evolution (Boore and Brown Citation1998; Curole and Kocher Citation1999; Boore et al Citation2005; Shen et al Citation2009). Hence, the complete mitochondrial genome sequence of X. revulsus was sequenced, assembled and characterized, which could provide important genetic data for studying of population genetics, molecular phylogenetics and evolutionary relationship of X. revulsus. All these provide important basic information for the conservation and utilization of this biome resource.
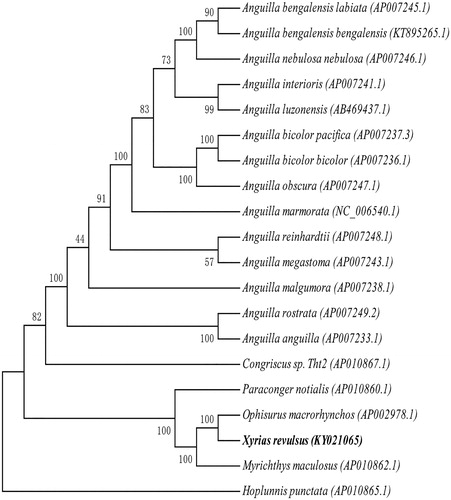
Total genomic DNA was isolated from each species using approximately of muscle tissue. Total DNA was eluted in sterile deionized water and was stored at −20 °C. The specimen was collected at the China-Vietnam Collective Fishery Zone (107.634 N, 19.714E) and stored at the Herbarium of Ocean College in Beibu Gulf University (X.R.001). Paired-end library (450 bp) was sequenced using Illumina Hiseq4000 platform, with 150 bp pair-end sequencing method. The mitochondrial genome was assembled using the chloroplast and mitochondrion assemble (CMA)V1.1.1 software (Guangzhou SCGene Co., Ltd), which was based on sequencing reads’ overlap and paired-end relationship. Protein-coding genes and rRNA genes were annotated with blast+(2.5.0) with allied species, and tRNAs were predicted with tRNAscan-SE v2.0 (http://lowelab.ucsc.edu/tRNAscan-SE/) (Lowe and Chan Citation2016). 11,226 raw reads with an average length of 150 bases and 1,683,900 nt were obtained, with average reads depth of 94.7X. Our research findings revealed that the circular genome is 17,784 bp, which consists of 13 protein-coding genes(PCGs), 2 D-loop, 2 rRNAs genes, 22 tRNAs genes, showing that the gene composition and arrangement are very close to the reported Ophisurus macrorhynchos composition (Shen et al. Citation2014). The gene composition of eels mitochondrial genomes is conservative.The contents of A, T, G, and C in mitochondrial genome were 32.45%, 25.76%, 15.72%, and 26.08%, respectively. An overall GC content of whole mitochondrial genome is 41.8%. The sequence was deposited in the GenBank (GenBank: KY021065). A phylogenetic analysis was conducted on 20 mitochondrial genomes. Maximum likelihood (ML) method was used for phylogenetic analysis. The best-fit models of evolution for the coding genes was selected by jmodeltest2 (https://github.com/ddarriba/jmodeltest2) (Darriba et al. Citation2012). RAxML v8.0.0 (RAxML Version 8 Citation2014) was used to build the tree with 1000 bootstrap (). Polygenetic analysis showed that the X. revulsus was very close to O. macrorhynchos. The complete mitochondrial genome of X. revulsus provided important genetic information for understanding phylogenetic relationships of Prosobranchia mitochondrial genome.
Figure 1. Phylogenetic relationships of Prosobranchia based on 20 mitochondrial genomes using NJ method. GenBank accession numbers: Anguilla bengalensis labiata (AP007245.1), Anguilla bengalensis bengalensis (KT895265.1), Anguilla nebulosa nebulosa (AP007246.1), Anguilla interioris (AP007241.1), Anguilla luzonensis (AB469437.1), Anguilla bicolor pacifica (AP007237.3), Anguilla bicolor bicolor (AP007236.1), Anguilla obscura (AP007247.1), Anguilla marmorata (NC_006540.1), Anguilla reinhardtii (AP007248.1), Anguilla megastoma (AP007243.1), Anguilla malgumora (AP007238.1), Anguilla rostrata (AP007249.2), Anguilla anguilla (AP007233.1), Congriscus sp. Tht2 (AP010867.1), Paraconger notialis (AP010860.1), Ophisurus macrorhynchos (AP002978.1), Xyrias revulsus (KY021065.1), Myrichthys maculosus (AP010862.1), Hoplunnis punctate (AP010865.1).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Our data is shared in ethically correct circumstances, without violating human protection, or other valid ethical, privacy, or security issues. The data that support the findings of this study are openly available in NCBI (https://www.ncbi.nlm.nih.gov/nuccore/KY021065.1/).
Additional information
Funding
References
- Anonymous. 1996. Fish of this month, Xyrias revulsus. Izu Oceanic Park Diving News, 8, 1. (In Japanese.)
- Boore JL, Brown WM. 1998. Big trees from litle genomes: mitochondrial gene order as a phylogenetic tool. Curr Opin Genet Dev. 8(6):668–674.
- Boore JL, Macey JR, Medina M. 2005. Sequencing and comparing whole mitochondrial genomes of animals. Meth Enzymol. 395:311–348.
- Curole JP, Kocher TD. 1999. Mitogenomics digging deeper with complete mitochondrial genomes. Trends Ecol Evol (Amst). 14(10):394–339.
- Chen WL. 2006. Preliminary studies on the taxonomy of snake-eel (Ophichthidae, Anguilliformes) off Eastern Taiwan. Taiwan, National Taiwan Ocean University.
- Darriba D, Taboada GL, Doallo R, Posada D, et a1. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Huang KQ, Zhan HX. 1981. New records of Chinese fishes collected from the East China Sea. Zool Systemat. 6(1):30.
- Jordan DS, Snyder JO. 1901. A review of the apodal fishes or eels of Japan, with descriptions of nineteen new species. Proc US Natl Museum. 23(1239):837–890.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Mccosker JE. 1998. Snake-eels of the genus Xyrias (Anguilliformes: Ophichthidae). Cybium. 22(1):7–13.
- Mccosker JE, Chen WL, Chen HM. 2009. Comments on the snake-eel genus Xyrias (Anguilliformes: Ophichthidae) with the description of a new species. Zootaxa. 2289(1):61–67.
- RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics; 2014.
- Shen X, Ma X, Ren J, Zhao F. 2009. A close phylogenetic relationship between Sipuncula and Annelida evidenced from the complete mitochondrial genome sequence of Phascolosoma esculenta. BMC Genom. 10(1):136–136.
- Shen X, Tian M, Meng XP, et al. 2014. Eels mitochondrial protein-coding genes translocation and phylogenetic relationship analyses. Acta Oceanol Sin (in Chinese). 36(4):73–81.
- Tan HJ. New records of fishes from China Sea. Investitgatio et Studium Naturae. 1983:25–26. (In Chinese.)
