Abstract
We newly obtained the near complete mitochondrial genome of Oreolalax schmidti (18,481bp) by using polymerase chain reaction (PCR) in this study. It includes 13 protein-coding genes (PCG), two ribosomal RNA (rRNA) genes, and 23 transfer RNA (tRNA) genes. The phylogenetic tree indicates that the Oreolalax schmidti is closely related to the O. lichuanensis. This report will help the further studies of Oreolalax species classification and source protection.
The Oreolalax schmidti belongs to the family Megophryidae, genus Oreolalax, which is endemic to China (Fei et al. Citation2012; Frost Citation2020) and distributed in Central Sichuan (Baoxing, Dujiangyan, Mount Emei, Hongya, Mianning, Shimian, and Wenchuan counties). This species lives in the mountain, regions at an altitude of 1700-2400 m, residing in bushes near small streams, in damp caves or under rocks in the streams, currently suffering from habitat loss (AmphibiaChina Citation2020). However, there is only a small portion of its mitochondrial genomes of O. schmidti available in the GenBank, e.g., 12 s rRNA and 16 s rRNA genes. This study along with the first near complete mitochondrial genome of O. schmidti may help us understand the information and phylogeny of this species.
During the field trip in 2015, the sample of O. schmidti was collected from Wawushan Nature Reserve (29°30′28.32″N, 102°51′53.20″E., elev. 2071 m), Sichuan Province, China, and the tissue was preserved in 99.9% ethanol. The specimen was deposited at the Museum of Sichuan Agricultural University (Specimen voucher: 20120281). Genomic DNA was extracted from leg muscle tissue by using the Ezup pillar genomic DNA extraction kit (Sangon Biotech, Shanghai, China). Finally, we sent DNA sample to Personal Biotechnology (Shanghai, China) for library construction and sequencing on an Illumina MiSeq platform (PE400).
The near complete mitogenome (MT773151) of O. schmidti is 18,481bp in length, which contained 38 genes including 13 protein coding genes (PCGs), two ribosomal RNA (rRNA) genes, 23 tRNA genes (including two trnM genes) and a partial control region sequence (about 1,498bp). The majority mitogenome length of O. schmidti is 18,481, and the base composition of the mitogenome is 28.3% A, 14.4% G, 24.5% C and 32.8% T, respectively. Comparative analysis indicates that the mitogenome structure of O. schmidti is similar to that of other Megophryidae (Xiang Citation2013; Liang Citation2016; Liang et al. Citation2016; Huang et al. Citation2020). Nad6 and eight tRNA (tRNA-Gln, Ala, Asn, Cys, Tyr, Ser, Glu, and Pro) are located on the L-light strand (L strand), and the others are encoded on the heavy strand (H strand). Most of the PCGS (nine) were started by ATG, except three PCGS (cox1, nad5, and nad6) by GTG, and nad3 by ATT. Cox1 was terminated by TAG, two PCGS (nad5 and nad6) terminated by AGG as termination codon, and four (nad2, atp8, nad3 and nad4l) terminated by TAA. Four PCGS (nad1, atp6, cox3 and nad4) stopped with TA, whereas two (cox2 and cytb) stopped with a single base T. The length of 12S and 16S rRNA are 938 and 1,581 bp respectively.
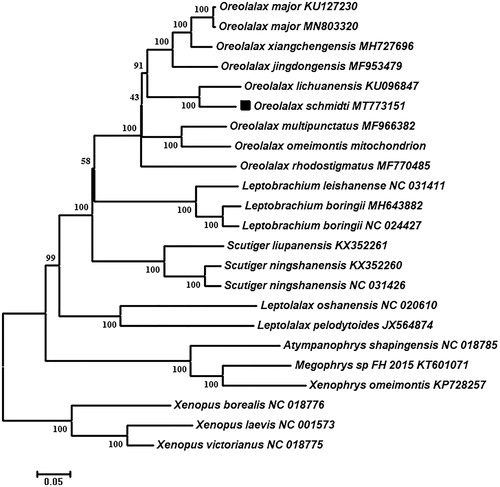
In order to determine the taxonomic status of O. schmidti, we constructed the phylogenetic tree by MEGA7.0 (Kumar et al. Citation2016) based on 13 PCGs. The final alignment consisted of 23 sequences of 20 species from eight genera (Oreolalax, Leptobrachium, Scutiger, Leptolalax, Atympanophrys, Megophrys, Xenophrys, and Xenopus), and we select three Xenopus species as outgroups. The phylogenetic trees indicated O. schmidti has the closest relationship with O. lichuanensis, and a monophyletic clade of genus Oreolalax were recovered and well sported with 100 percent bootstrap support (). This near complete mitogenome would contribute to further investigations of molecular evolution and conservation of genus Oreolalax.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data used to support the findings of this study are available in GenBank at https://www.ncbi.nlm.nih.gov/nuccore/ MT773151, reference number MT773151.
References
- AmphibiaChina. 2020. The database of Chinese amphibians. Kunming Institute of Zoology (CAS). Kunming, Yunnan, China. http://www.amphibiachina.org/.
- Fei L, Ye C, Jiang J. 2012. Colored Atlas of Chinese amphibians and their distributions. Chengdu: Sichuan Publishing House of Science and Technology.
- Frost D. 2020. Amphibian species of the world: an online reference. Version 6.0. American Museum of Natural History, New York, USA. http://research.amnh.org/herpetology/amphibia/index.html.
- Huang T, Cui L, Li D, Fan X, Yang M, Yang D, Ni Q, Li Y, Yao Y, Xu H, et al. 2020. The complete mitogenome of the large toothed toad, Oreolalax major (Anura: Megophryidae) with phylogenetic analysis. Mitochondr DNA Part B. 5(1):1117–1118.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Liang X. 2016. Structure and evolution of mitochondrial genomes of megophryidae [PhD Dissertation]. Beijing: University of Chinese Academy of Sciences.
- Liang X, Wang B, Li C, Xiang T, Jiang J, Xie F. 2016. The complete mitochondrial genome of Oreolalax major (Anura: Megophryidae). Mitochondr DNA Part B. 1(1):118–119.
- Xiang T. 2013. Analysis of complete mitochondrial genomes in representative species of three subfamilies of Megophryidae (Amphibia, Anura) [MsD Dissertation]. Beijing: University of Chinese Academy of Sciences.