Abstract
In this paper, we characterized the mitogenomes of three ‘eyed’ turtles, the Beal’s Eyed turtle (Sacalia bealei), the Four Eye-spotted turtle (S. quadriocellata), and the Hainan Four Eye-spotted turtle which was a unique lineage in the four-eyed turtle and considered as an independent species S. insulensis recently. The full lengths of the S. bealei and S. quadriocellata mitogenomes are 16,564 bp, and 16,555 bp, respectively, while the length of partial mitochondrial genome of S. insulensis is 16,433 bp without tailed part of D-loop. All the genes exhibit the typical mitochondrial gene arrangement and transcribing directions of turtles. Phylogenetic analysis indicating that a deep divergence about 7.8% p distance was found between S. bealei and (S. quadriocellata + S. insulensis), and the divergence (2.8% in patristic distance) between S. quadriocellata and S. insulensis is comparable with other closely related species in turtles.
Turtles of genus Sacalia are currently composed of two species: the Beal’s Eyed turtle (S. bealei) and the Four Eye-spotted turtle (S. quadriocellata). Sacalia bealei is endemic to southern China, while the distribution of S. quadriocellata ranges from Vietnam and Laos in southeast Asia to southern China and Hainan island (Turtle Taxonomy Working Group Citation2017). Due to over-harvesting for pet trade, they have become extremely rare in the wild (Hu et al. Citation2016; Gong et al. Citation2017) and listed as endangered on the IUCN Red List (Rhodin et al. Citation2018) .
Sacalia quadriocellata from Hainan island is unique in appearance compared to other populations from mainland China and Southeast Asia. Previous studies based on phylogenetic analysis of CytB gene, and comparative morphological analysis indicated that it should be classified as a valid species named as the Hainan Four Eye-spotted turtle (S. insulensis; Shi et al. Citation2008; Lin et al. Citation2018). This suggestion has been accepted in the latest updated checklists of reptiles of China (Wang et al. Citation2020).
In this study, we characterized and compared the complete mitogenomes of the three ‘eyed’ turtles collected from southern China, with the S. bealei from Zhangzhou (117.37 E, 24.53 N), Fujian province, the S. quadriocellata from Yangjiang (111.59 E, 22.21 N), Guangdong province and the S. insulensis from Qiongzhong (109.88E, 19.06N), Hainan island. They were wild caught and then bred alive in captive at Turtles Research and Conservation Center of Hainan Normal University, Hainan Province, China (110.35E, 20.00N). Blood samples were collected from the sub-vertebral site of live turtles following the McArthur et al. (Citation2004) methodology, and stored at this center with code EyedTur01, EyedTur02 and EyedTur03. Genomic DNA was extracted with the DNeasy Blood and Tissue Mini Kit (QIAGEN, Hilden, Germany), following the product protocol. 7.0–7.2 Gb bases were sequenced for each sample, in 150 bp each read, pair end, with the Illumina NovaSeq 6000 System. Raw reads were trimmed for Illumina adapters at right end and low-quality bases at both ends with BBDuk Trimmer 1.0 and mapped to public mitogenome sequences S. bealei (GenBank Accession Number GU183364) and S. quadriocellata (GU320209), with Low Sensitivity/Fastest, Fine Tuning(None), in Geneious Prime® 2020.0.3. 14,296, 11,303 and 1820 reads were matched to the reference respectively for S. bealei, S. quadriocellata and S. insulensis, generating contigs with an average coverage at 128.4, 101.6 and 16.4-fold. Consensus sequences were generated for bases covered with at least 2 reads and annotated according to the references with the Transfer Annotations function in Geneious Prime® 2020.0.3, which were accessible in GenBank with accession numbers MT372820–MT372822.
The length of complete mitochondrial genome of S. bealei and S. quadriocellata is 16,564 bp and 16,555 bp, and the length of partial mitochondrial genome of S. insulensis is 16,433 bp without tailed part of D-loop. The structure of mitochondrial genomes of the three ‘eyed’ turtles are similar to other turtle species, including 13 protein coding genes, 2 rRNA genes, 22 tRNA genes, and 1 control region, whose base composition is 34% A, 26% C, 13% G, 27% T, and 39% GC. Most genes are coded in heavy strand except tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Glu, tRNA-Pro and ND6 gene. Most protein-coding genes use common start codon ATG and stop codon TAA, except rare start codon GTG and rare stop codon AGG in CO1 gene. It is common that TAA stop codon is completed by the addition of 3′-A residues to the mRNA.
Mitogenome sequences of closely related species were aligned with our sequences by Mafft Plugin 1.4.0 installed in Geneious Prime. The phylogeny was inferred from 15,590 bp coding regions excluding the D-loop and nearby tRNA-Phe, using the Bayesian inference in MrBayes plugin 2.2.3 (Huelsenbeck and Ronquist Citation2001; Ronquist and Huelsenbeck Citation2003) installed in Geneious Prime, with GTR + I + G suggested by Modeltest (Posada and Crandall Citation1998), and 1,100,000 iterations with beginning 100,000 burn-in were performed.
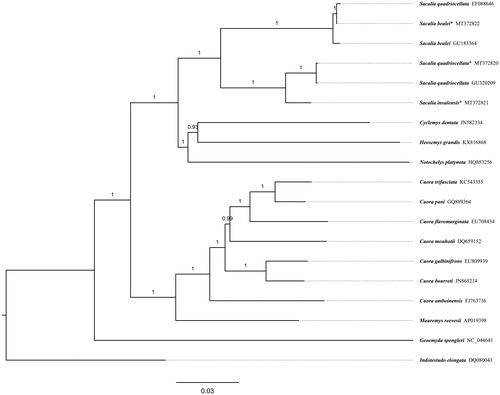
In , all sequences from Genus Sacalia were clustered into a monophyletic group located in the Family Geoemydidae. A deep divergence about 7.8% p distance was found between S. bealei and (S. quadriocellata + S. insulensis). There is perhaps an identification error that sequence EF088646 is labeled as S. quadriocellata in GenBank but is almost exactly the same as the two sequences from S. bealei, sharing more than 99% identical sites each other. The divergence between S. quadriocellata and S. insulensis is about 2.8% in patristic distance, comparable to that between closely related species in genus Cuora, such as 3.0% between C. trifaciata and C. pani, and 3.8% between C. galbinifrons and C. bourreti. The results obtained here can contribute to phylogenetic analysis and biological conservation of ‘eyed’ turtles further.
Figure 1. Bayesian tree of ‘eyed’ turtles Sacalia spp. based on mitogenomes excluding D-loop and nearby tRNA-Phe. Indotestudo elongata (DQ080043) was set as an outgroup. Posterior probability values are shown at branches. Scientific names and GenBank accessions number are labeled at tips. Mitogenomes sequenced in this study are marked with an asterisk.
Acknowledgement
This work was approved by Animal Research Ethics Committee of Hainan Provincial Education Center for Ecology and Environment, Hainan Normal University (HNECEE-2014-001).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT372820, MT372821 and MT372822.
Additional information
Funding
References
- Gong SP, Shi HT, Jiang AW, Fong JJ, Gaillard D, Wang JC. 2017. Disappearance of endangered turtles within China’s nature reserves. Curr Biol. 27(5):170–171.
- Hu QR, Yang JB, Lin L, Xiao FR, Wang JC, Shi HT. 2016. Habitat selection factors of the Beal′s Eyed Turtle (Sacalia bealei) at Huboliao National Nature Reserve, Fujian Province. Chin J Zool. 51(4):517–528. (in Chinese).
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Lin L, Sun L, Wang W, Shi HT. 2018. Taxonomic status and nomenclature of four eye-spotted turtle from Hainan Island. Sichuan J Zool. 37(4):435–438. (in Chinese).
- McArthur S, Wilkinson R, Meyer J. 2004. Medicine and surgery of tortoises and turtles. Oxford, UK: Blackwell Publishing Ltd.
- Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics. 14(9):817–818.
- Rhodin AGJ, Stanford CB, Dijk PPV, Eisemberg C, Luiselli L, Mittermeier RA, Hudson R, Horne BD, Goode EV, Kuchling G, et al. 2018. Global conservation status of turtles and tortoises (Order Testudines). Chelonian Conserv Biol. 17(2):135–161.
- Ronquist F, Huelsenbeck JP. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Shi H, Fong JJ, Parham JF, Pang J, Wang J, Hong M, Zhang Y-P. 2008. Mitochondrial variation of the “eyed” turtles (Sacalia) based on known-locality and trade specimens. Mol Phylogenet Evol. 49(3):1025–1029.
- Turtle Taxonomy Working Group. 2017. Turtles of the world: annotated checklist and atlas of taxonomy, synonymy, distribution, and conservation status. In: Rhodin AGJ, Iverson JB, van Dijk PP, Saumure RA, Buhlmann KA, Pritchard PCH, Mittermeier RA, editors. Conservation biology of freshwater turtles and tortoises: a compilation project of the IUCN/SSC tortoise and freshwater turtle specialist group. Chelonian research monograph 8th edn, Vol. 7. Lunenburg, MA; New York, NY: Chelonian Research Foundation; Turtle Conservancy; p. 1–292.
- Wang K, Ren JL, Chen HM, Lyu ZT, Guo XG, Jiang k, et al. 2020. The updated checklists of amphibians and reptiles of China. Biodivers Sci. 28(2):189–218. (in Chinese)
