Abstract
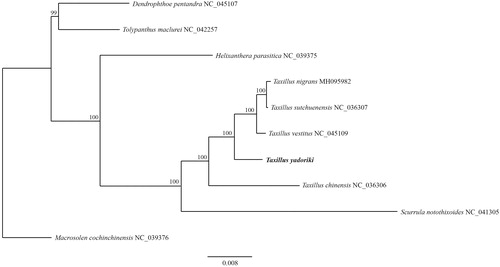
Taxillus yadoriki (Loranthaceae) is a hemiparasitic evergreen shrub distributed in Korea and Japan. We report the complete chloroplast genome of T. yadoriki to provide insight into the phylogenetic relationship of Loranthaceae. This genome is 122,192 bp long, with two IR regions (22,756 bp each) that separate a large single-copy (LSC) region (70,628 bp) and a small single-copy (SSC) region (6052 bp). It contains 109 genes that encode 68 proteins, 8 rRNAs, and 33 tRNAs. All of ndh genes have been lost and the SSC region consists of only four genes similar to other Taxillus species. In ML phylogenetic, monophyly of Taxillus was strongly supported with high bootstrap value and formed a sister group with Scurrula.
Taxillus L., comprises approximately 30 species that are widely distributed, with the majority occurring on the warm-temperate and subtropical regions in Asia (Wu et al. Citation2003). The genus displays a lifestyle as hemiparasites, specialized by parasitic organs called haustoria (Li et al. Citation2017). The chloroplast genome (cp) of Taxillus has undergone structural changes and gene loss that play a key role in the evolution from autotrophy to parasitism (Li et al. Citation2017; Zhao et al. Citation2019). However, to date, only a few plastid genome (plastome) has been sequenced for Taxillus.
Taxillus yadoriki (Maxim.) Danser (Loranthaceae) is a hemiparasitic evergreen shrub distributed in Korea and Japan (Ohba Citation2006). Also, the species is a very important medical plant with various pharmacological activities such as anti-cancer, anti-inflammation (Park et al. Citation2018). Thus, we characterized the chloroplast genome of T. yadoriki and analyzed its phylogenetic position within Loranthaceae.
In this study, material of T. yadoriki were collected from the Jeju Island in South Korea (N 33°15′23", E 126°37′23"). The voucher specimen (Lee. 2005039) was stored at the herbarium in the Department of Biology Education, Chonnam National University (BEC). The DNA library was constructed and sequenced using MGI-seq 2000 platform (LAS, Seoul, Korea), generating 34,340,232 raw reads (150 bp paired-end). After trimming the sequences, the obtained clean data were mapped with the reference cp genome for T. sutchunensis (NC.036037), using Geneious 10.2.3 (Kearse et al. Citation2012). On that genome, 3,926,088 reads were assembled with an average of 4808X coverage (max: 6916X, min: 1146X). The annotation was separately performed using DOGMA (Wyman et al. Citation2004), and were manually corrected for start and stop codons and for intro/exon boundaries. The annotated cp genome sequence was deposited in the GenBank with Accession Number MT702883. To construct the phylogenetic tree, we downloaded complete cp genome sequences of 9 related species from the NCBI database and aligned using the MAFFT (Katoh and Toh Citation2010). We performed maximum likelihood (ML) analysis with RAxML v.8.0 (Stamatakis Citation2014) using default parameters and 1000 bootstrap replicates.
The cp genome structure of T. yadoriki is 122,192 bp long, with 2 IR regions (22,756 bp each) that separate a large single-copy (LSC) region (70,628 bp) and a small single-copy (SSC) region (6,052 bp). Overall G + C content of the genome is 37.3%, compared with 34.6% for the LSC and 42.8% for the IRs. This cp genome contains 109 genes that encode 68 proteins, 8 rRNAs, and 33 tRNAs. All of the ndh genes have been lost and the SSC region consists of only four genes (trnLUAG, ccsA, psaC, ycf1) similar to other Taxillus species (Li et al. Citation2017). The trnLUAG was located on the IRA and SSC region border similar to other Loranthaceae species. In ML phylogenetic, monophyly of Taxillus was strongly supported with high bootstrap value (100%) and formed a sister group with Scurrula (). Taxillus yadoriki is sister to a clade consisting of T. nigrans, T. vestitus and T. sutchuenensis. These new phylogenetic data provide insight into the evolutionary progress of Loranthaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI (accession no. MT702883) at https://www.ncbi.nlm.nih.gov.
Additional information
Funding
References
- Katoh K, Toh H. 2010. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 26(15):1899–1900.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li Y, Zhou JG, Chen XL, Cui YX, Xu ZC, Li YH, Song JY, Duan BZ, Yao H. 2017. Gene losses and partial deletion of small single-copy regions of the chloroplast genomes of two hemiparasitic Taxillus species. Sci Rep. 7(1):12.
- Ohba H. 2006. Loranthaceae. In: Iwatsuki K, Boufford DE, Ohba H, editors. Flora of Japan. Vol. IIa. Tokyo: Kodansha; p. 117–119.
- Park SB, Park GH, Kim HN, Song HM, Son HJ, Park JA, Kim HS, Jeong JB. 2018. Ethanol extracts from the branch of Taxillus yadoriki parasitic to Neolitsea sericea induces cyclin D1 proteasomal degradation through cyclin D1 nuclear export. BMC Complement Altern Med. 18(1):189.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wu Z, Raven PH, Hong D. 2003. Loranthaceae. In: Huaxing Q, Gilbert MG, editors. Flora of China. Vol. 5. Beijing: Science Press; p. 231–238.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Zhao S, Li J, Ma R, Miao N, Mao Q, Mao K. 2019. Characterization of the complete chloroplast genome of Taxillus nigrans. Mitochondrial DNA Part B. 4(1):472–473.