Abstract
Symphoricarpos sinensis is an important ornamental plant in China. Due to the lack of efficient molecular markers, although recent studies supported that Symphoricarpos was phylogenetically closely related to Lonicera, Triosteum, Heptacodium, and Leycesteria, relationships among these taxa remained uncertain. To examine the phylogenetic position of Symphoricarpos within Caprifoliaceae, we characterized the complete chloroplast (cp) genome of S. sinensis. The results showed that S. sinensis shared a typical quadripartite structure as with most angiosperms. It was 155,738 bp in length, including two inverted repeat (IR) regions (24,010 bp), a small single-copy (SSC) region (18,942 bp) and a large single-copy (LSC) region (88,776 bp). Phylogenetic analysis demonstrated that Caprifoliaceae and Adoxaceae formed two distinct monophyletic clades, and S. sinensis was closely related to lonicera and Triosteum, albeit with low support. The whole cp genome of S. sinensis will be useful resources for future studies on phylogeny and conservation in Symphoricarpos.
Symphoricarpos (Caprifoliaceae) is an upright shrub distributed in North America, Mexico and China. Based on recent molecular phylogenetic studies, Symphoricarpos was found to be closely related to Lonicera, Triosteum, Heptacodium, and Leycesteria, however, phylogenetic relationships among these taxa remained controversial (Xiang et al. Citation2020). In recent years, the whole cp genomes have become valuable resources for molecular phylogeny and species identification due to highly conservative structure and low evolutionary rate (Liu et al. Citation2017; Liang et al. Citation2018). In this study, we reported the complete cp genome of S. sinensis, which will be helpful for further studies on phylogeny in Symphoricarpos.
Fresh materials were collected in Zhongshan Botanical Garden (N 32°03'16.85″, E 118°49′52.06″) and the voucher specimen was stored in Henan Agricultural University Herbarium (CYN2019052202). The whole cp genome of S. sinensis was sequenced on an Illumina Hiseq2500 platform at Suzhou Jinweizhi Biotechnology Institute. Firstly, the filtration and assembly of chloroplast genomes was completed at CLC Genomics Workbench (http://www.clcbio.com). Then, the cp genome of S. sinensis was annotated in PGA (http://github.com/quxiaojian/PGA) using the Lonicera japonica (NC_026839.1), Heptacodium miconioides (NC_042739) and Triosteum pinnatifidum (NC_037952) as references. The start/stop codons and intron/exon boundaries of genes were subsequently manually modified based on the reference sequences. Next, tRNAscan-SE v1.21 (Schattner et al. Citation2005) was further used to examine the tRNA boundaries with default settings, and then the physical map of the cp genome was drawn using the online program Organellar Genome DRAW (Lohse et al. Citation2013). Finally, to infer the phylogenetic position of Symphoricarpos within Caprifoliaceae, 15 species of Lonicera and 8 species representing 8 genera of Caprifoliaceae were selected to construct the phylogenetic tree using PhyML v3.0 (Guindon et al. Citation2010). Six species (i.e. Adoxa moschatellina, Tetradoxa omeiensis, Sinadoxa corydalifolia, Viburnum utile, Viburnum betulifolium and Sambucus williamsii) were selected as outgroups.
The cp genome of S. sinensis (GenBank accession number: MT507606) harbored a typical quadripartite structure with a full length of 155,738 bp and 38.4% GC content, comprising two inverted repeat (IR) regions (24,010 bp), a LSC region (88,776 bp) and a SSC region (18,942 bp). It encoded 131 functional genes, including 84 protein-coding genes, 39 transfer RNA (tRNA) genes, 8 ribosomal RNA (rRNA) genes. Of those protein-coding genes, 9 contained one intron and 4 contained two introns. It was worth noting that 16 genes were completely duplicated in the IR regions. The overall structure, gene content and arrangement of the cp genome of S. sinensis was quite similar to those of other reported Caprifoliaceae species (He et al. Citation2017; Wang et al. Citation2020).
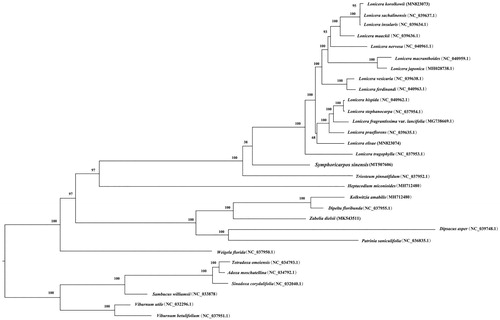
The phylogenetic analysis () recovered two well-supported monophyletic clades (both 100% bootstrap values) representing Caprifoliaceae and Adoxaceae, respectively, and showed that S. sinensis was closely related to lonicera and Triosteum within Caprifoliaceae, albeit with low bootstrap support (38%). These results were largely consistent with previous studies (Bell Citation2010; Xiang et al. Citation2020).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/WebSub/?form=history&tool=genbank, reference number [MT507606].
Additional information
Funding
References
- Bell C. 2010. Towards a species level phylogeny of Symphoricarpos (Caprifoliaceae), based on nuclear and chloroplast DNA. Syst Bot. 35(2):442–450.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyMl 3.0. Syst Biol. 59(3):307–321.
- He L, Qian J, Li XW, Sun ZY, Xu XL, Chen SL. 2017. Complete chloroplast genome of medicinal plant Lonicera japonica: genome rearrangement, intron gain and loss, and implications for phylogenetic studies. Molecules. 22(2):249.
- Liang FP, Wen XN, Gao HY, Zhang Y. 2018. Analysis of chloroplast genomes features of Asteraceae species. Genomics Appl Biol. 37:5437–5447.
- Liu LX, Li R, Worth JRP, Li X, Li P, Cameron KM, Fu CX. 2017. The complete chloroplast genome of Chinese Bayberry (Morella rubra, Myricaceae): implications for understanding the evolution of Fagales. Front Plant Sci. 8:968.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellargenome DRAW: a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:575–581.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:686–689.
- Wang HX, Liu H, Moore MJ, Landrein S, Liu B, Zhu ZX, Wang HF. 2020. Plastid phylogenomic insights into the evolution of the Caprifoliaceae s.l. (Dipsacales). Mol Phylogenet Evol. 142:106641.
- Xiang CL, Dong HJ, Landrein S, Zhao F, Yu WB, Soltis DE, Soltis PS, Backlund A, Wang HF, Li DZ, et al. 2020. Revisiting the phylogeny of Dipsacales: new insights from phylogenomic analyses of complete plastomic sequences. J Syst Evol. 58(2):103–117.