Abstract
Rumex nepalensis is a traditional Chinese herb used for detoxification, hemostasis, and anti-inflammatory. In this study, the complete chloroplast sequence of R. nepalensis was characterized from Illumina pair-end sequencing. The cpDNA was 159,044 bp in length, containing a pair of inverted repeats of 30,623 bp each separated by a large and small single copy region of 84,819 bp and 12,979 bp, respectively. The complete chloroplast genome of R. nepalensis contained 128 genes, 37 tRNA, and 8 rRNA. Phylogenetic tree demonstrated that R. nepalensis was closely related to Rumex acetosa.
Rumex nepalensis (R. nepalensis) Spreng. belongs to the Rumex L, Polygonaceae family. It is a traditional Chinese herb employed for detoxification, hemostasis, and anti-inflammatory properties (Li et al. Citation1998). At present, there are few studies on the R. nepalensis genome. Here, we assembled and characterized the complete chloroplast genome sequence of R. nepalensis to provide information for the identification of Rumex L, as well as assist further phylogenetic study of Polygonaceae. The annotated genome sequence has been deposited into Genbank with the accession number MT323235.
The leaves of R. nepalensis were collected from Nyingchi (Tibet, China; N: 29°39′ 16.29″, E: 94°18′ 04.07″). The specimens of R. nepalensis have been kept in Tibet Agriculture & Animal Husbandry University. The specimen accession number is 542621150709007LY. Total genomic DNA of R. nepalensis was isolated by a modified CTAB method (Allen et al., Citation2006) and sequenced as the paired end using the Illumina HiSeq 2000 platform (Illumina, San Diego, CA). A total of 1.9 G base pairs of sequencing data were obtained and then de novo assembled using the CLC genome assembler program (ver.4.06 beta, CLC Inc, Aarhus, Denmark) as previously described (Kim et al. Citation2015). The Geneious Prime (Kearse et al. Citation2012) was used to assemble the complete chloroplast genome, with Rheum palmatum (GenBank: NC027728) as the reference. DOGMA program was used for annotation of the complete chloroplast genome (Wyman et al. Citation2004) and the annotation was corrected with Geneious Prime (Kearse et al. Citation2012).
The complete R. nepalensis chloroplast genome is a circular molecular structure of 159,044 bp in length with 37.6% GC content, consisting of two inverted repeats (30,623 bp) separated by a large single-copy region (84,819 bp) and a small single-copy region (12,979 bp). The complete chloroplast genome of R. nepalensis contained 128 genes, 37 tRNA and 8 rRNA.
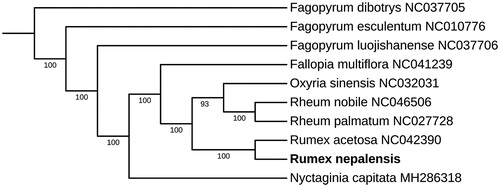
To investigate the phylogenetic relationship of R. nepalensis, the phylogenetic tree was generated by the maximum-likelihood (ML) method from alignments created using the MAFFT (Katoh and Standley Citation2013) and trimmed properly by trimAl (Capella-Gutierrez et al. Citation2009). We selected nine published Polygonaceae complete chloroplast genomes from GenBank to assess the genetic and phylogenetic relationship with R. nepalensis. The ML tree was constructed using IQ-TREE (Lam-Tung et al. Citation2015) with 1000 replicates. As shown in the phylogenetic tree (), R. nepalensis is most closely related to R. acetosa. The result was consistent with the traditional plant morphological taxonomy. The complete chloroplast genome sequence of R. nepalensis facilitates the phylogenetic studies of Polygonaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/, reference number MT323235.
Additional information
Funding
References
- Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF. 2006. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc. 1(5):2320–2325.
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kim K, Lee S-C, Lee J, Yu Y, Yang K, Choi B-S, Koh H-J, Waminal NE, Choi H-I, Kim N-H, et al. 2015. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655.
- Lam-Tung N, Schmidt HA, Arndt VH, Quang MB. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Li AR. 1998. The flora of China. Beijing: Science Press, 25, 160-16.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.