Abstract
The DNA barcode data of Asian Glass Lizard, Dopasia gracilis, is limited in the global database, especially from India. The present study aimed to generate a barcode sequence of morphologically identified D. gracilis from the Mizoram state in northeast India and compared with other Anguidae species. The studied species showed monophyletic clustering in the Bayesian analysis (BA) phylogeny with strong posterior probability support and also discriminated sufficient Kimura 2 parameter genetic distances. The barcode data of D. gracilis revealed high intra-species genetic variability and formed two clusters in BA phylogeny. The Templeton, Crandall, and Sing network also depicted four different haplotypes within the barcode sequences of D. gracilis. The DNA sequences generated from northeast India showed 6.5–6.6% and 7.3% genetic distances with the sequences generated from Yunnan Province and Tibetan Plateau, respectively. Considering the high genetic distances, multiple clustering, and distinct haplotypes, the present study assumed the presence of possible cryptic diversity of D. gracilis in the Indochina sub-region and a distinct population in northeast India. We recommended the generation of more DNA information from different localities to elucidate the actual diversity of D. gracilis within the known range distribution.
Introduction
The members of family Anguidae are carnivorous reptiles having a unique conserved body plan and either ground-dwelling or arboreal habits in nature (Lin et al. Citation2003). The Anguidae comprises 102 species within 15 genera and has widespread distribution in North, Central, and South America, West Indies, Europe, Asia, and North Africa (McConkey Citation1954; Pough et al. Citation1998). Among them, the subfamily Anguinae consists of about 20 described species (six are distributed in Europe, one in North Africa, five in Asia, two in Indonesia, and six in North America) (Lavin and Girman Citation2019). Due to the complex taxonomic characters and lack of keys, the classification of Anguinae species has been repeatedly revised (Gvoždík et al. Citation2010; Nguyen et al. Citation2011). For example, the genus Ophisaurus was believed to include 12 species, widely distributed in North to Central America, East and Southeast Asia, and North Africa. Later on, based on a taxonomic revision and subsequent conformity, the generic name of East and Southeast Asian Ophisaurus species was erected to the genus Dopasia (Brygooé Citation1987; Macey et al. Citation1999; Conrad Citation2008; Nguyen et al. Citation2009; Conrad et al. Citation2011). Currently, the genus Dopasia consists of seven species; five are distributed in the Indochina sub-region, one is distributed in Malaysia (Sabah, Sarawak), and Indonesia (Borneo), and one is endemic to Indonesia (Sumatra) (Nguyen et al. Citation2011; Uetz et al. Citation2020).
The limbless Asian Glass Lizard or Burmese Glass Lizard, Dopasia gracilis Gray, Citation1845 is originally described from the Khasi Hills in the state of Meghalaya, northeast India. This species can be distinguished based on the number of vertebrae from atlas to remnants of hind leg bones (Brygooé Citation1987; Nguyen et al. Citation2011). Dopasia gracilis is widely distributed in India, China, Myanmar, Laos, Thailand, and Vietnam (Nguyen et al. Citation2009, Citation2011). Because of the deforestation produced by agriculture and plantations and the exploitation of biological resources, this lizard group confronts threats of habitat destruction throughout their known distribution range including northeast India (IUCN Citation2020). Due to insufficient systematics studies, the conservation status of D. gracilis has not yet been assessed by the International Union for the Conservation of Nature (IUCN). Apart from taxonomic studies, few integrated approaches have been induced to understand this charismatic reptile group (Gvoždík et al. Citation2010, Citation2013; Pan et al. Citation2015; Song et al. Citation2015, Lavin and Girman Citation2019; Cai et al. Citation2020). The advancement of molecular tools is not only helpful to identify species, but is valuable information to reconstruct phylogenetic relationship (Wiens and Slingluff Citation2001; Vidal and Hedges Citation2005; Townsend et al. Citation2008; Wüster et al. Citation2008; Vidal and Hedges Citation2009; Wiens et al. Citation2010; Mulcahy et al. Citation2012; Pyron et al. Citation2013), to detect cryptic diversity (Laopichienpong et al. Citation2016), and to understand the origin as well as the group diversification (Sanmartín et al. Citation2001; Burbrink and Lawson Citation2007; Guo et al. Citation2012). In previous studies, both nuclear and mitochondrial genes were successfully used to discriminate Anguinae species, including D. gracilis (Lavin and Girman Citation2019). Besides phylogenetic relationships and divergence dating, potential cryptic diversity in D. gracilis with sufficient genetic variability was depicted sampling from China, Myanmar, and Vietnam. Owing to the wide biogeographic range distribution, this study was triggered with the hypothesis that the northeast Indian population of D. gracilis might be distinct, which warranted further confirmation through molecular studies. Also the DNA sequence of northeast Indian D. gracilis has never been compared with other generated sequences from their existing distribution. In this milieu, the present study is aimed to generate the DNA barcode data of D. gracilis from the Mizoram state and to compare these with other publicly available database sequences by estimating genetic distance, phylogenetic analysis, and haplotype network to check their genetic distinctiveness.
Materials and methods
The D. gracilis specimen was collected from the Mizoram state (23.78N 92.72E) in northeast India () and was morphologically identified based on the original description and previous literature, and with the help of naturalist’s guides (Gray Citation1845; Lalremsanga et al. Citation2010; Das and Das Citation2017). Field work and sampling were performed after obtaining prior permission (No.A.33011/2/99-CWLW/225) from the Chief Wildlife Warden of Environment, Forests and Climate Change, Govt. of Mizoram, India. Tissue sample was collected and preserved in 70% ethanol until downstream analysis. The entire specimen was preserved and vouchered (MZMU1329) in the Department of Zoology, Mizoram University, India. The molecular analyses were executed as per previously published protocols (Kundu et al. Citation2018). The genomic DNA was stored at the Center for DNA Taxonomy Laboratory, Molecular Systematics Division, Zoological Survey of India, Kolkata. The low-quality reads and gaps within the generated sequence were checked by SeqScanner V1.0 (Applied Biosystems Inc., CA, USA), nucleotide BLAST (https://blast.ncbi.nlm.nih.gov/), and ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/). The generated sequence was submitted to the GenBank database to acquire the accession number. The dataset was constructed by using 27 publicly available database sequences of nine Anguidae species. Two database sequences of Anniella geronimensis (EU445975) and Anniella pulchra (EU445968) under family Anniellidae were used as an out-group. The dataset was aligned by ClustalX (Thompson et al. Citation1997) and Kimura 2 parameter (K2P) genetic distances were calculated by MEGAX (Kumar et al. Citation2018). The suitable model for Bayesian analysis (BA) was estimated through MrModeltest v2 with lowest BIC value (Nylander Citation2004). The BA phylogeny was constructed in Mr. Bayes 3.1.2 by selecting nst = 6 and rates = invgamma for GTR + G + I model. The MCMC (one cold and three hot chains) was run for 1,000,000 generations with 25% burn-in and trees saving at each 100 generation (Ronquist and Huelsenbeck Citation2003). The phylogeny was further illustrated in web-based iTOL tool (https://itol.embl.de/) (Letunic and Bork Citation2007). The haplotype network was further constructed to infer the genealogical link within the different population of D. gracilis. The numbers of haplotypes, haplotype diversity (Hd), and number of polymorphic sites were calculated by using DnaSP v6 (Rozas et al. Citation2017). The haplotype network was constructed using PopART (http://popart.otago.ac.nz) (Leigh and Bryant Citation2015) with standard Templeton, Crandall, and Sing (TCS) method (Clement et al. Citation2000).
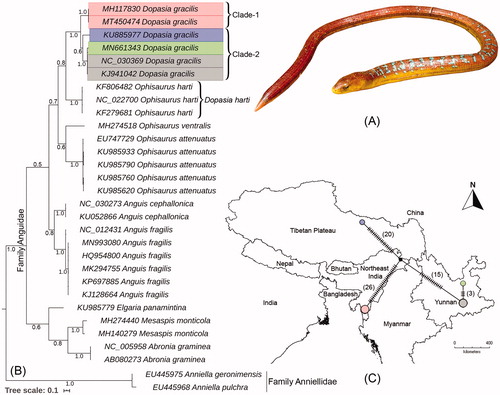
Figure 1. (A) Live photograph of D. gracilis collected from Mizoram state in northeast India, (B) Bayesian phylogeny based on partial mtCOI gene inferred the relationship of D. gracilis with other Anguidae species. (C) TCS network revealed distinct haplotype of D. gracilis in northeast India as compared with other Chinese population. The estimated haplotypes are shown in different colors vertex in collection locality map as used in phylogeny.
Results and discussion
Evolutionary diversity is associated with specific topography, climates, soil, and vegetation that stimulate an unparalleled distribution of higher level taxon around the world (Antão et al. Citation2020). Although the morphology of an organism from different geographical locations sometimes looks similar due to their static phenotypic characters and selection regime, their genetic constitution might have changed corresponding to their habitat, history, and environmental interaction. To explore this scenario, the interventions of molecular tools are a fruitful tool for taxonomy. Considering the abrupt losses of biodiversity due to climate and habitat changes, connect the earth observation through DNA sequences offering an efficient monitoring of biodiversity, their functions, and services (Bush et al. Citation2017). Considering the abrupt losses of biodiversity in recent past, the DNA sequences offers an efficient platform for illuminating the extant earth’s biota quickly and reliably (Bush et al. Citation2017). Since the inception of DNA barcoding, genetic traits are more widely used in evolutionary studies, an enormous nucleotide database with multiple species delimitation methods have proliferated rapidly for answering several biological questions throughout the globe (Nagy et al. Citation2012; Chambers and Hebert Citation2016). Noteworthy, the evolutionary relationships between and within species is crucial for biodiversity conservation planning, which has been recently illustrated for amphibian and reptile species occurring in a biodiversity hotspot region (Jablonski et al. Citation2016; Carvalho et al. Citation2017). Focusing on the widespread geographical distribution of the Asian Glass Lizard, the intra-species genetic information is still lacking from India. Hence, the present study compared the DNA barcode data of D. gracilis from northeast India and other geographical location to elucidate their intra-species genetic variability. The overall mean genetic distance of the Anguidae species was 19.4% in the present dataset. Excluding the singleton species, the mean intra-species genetic distance ranged from 0% (Anguis cephallonica, Abronia graminae, and Dopasia harti) to 5.1% (D. gracilis). The inter-species genetic distance ranged from 13.5% (A. cephallonica and Anguis fragilis) to 30.8% (A. graminae and D. harti). The BA phylogeny showed cohesive clustering of all studied species with high posterior probabilities support (). Dopasia gracilis showed sister relationship with D. harti (named as Ophisaurus harti in the database). Considering the high mean intra-species genetic distances (5.1%), we primarily depicted three clades (one clade from northeast India, one clade from Tibetan Plateau, China, and one clade from Yunnan Province, China). Further, due to the low posterior probability support (0.6) between two Chinese clades, we recognized two distinct clades (Clade-1 from India and Clade-2 from China) and assumed possible cryptic diversity of D. gracilis in Indochina sub-region. Altogether, the DNA barcode data of D. gracilis revealed four distinct combinations with 0.866 haplotype diversity and 61 polymorphic sites. For better illustration, the TCS haplotype network was also drawn and merged into the collection locality map (). The three database sequences of D. gracilis (collected from Mile and Gejiu City of Yunnan Province, China) showed 0.5% genetic distances with each other. However, the sequence generated from Tibetan Plateau, China, showed 5.5–5.8% genetic distance with the sequences generated from Yunnan Province, China. The barcode data generated from northeast India showed 6.5–6.6% and 7.3% genetic distances with the sequences generated from Yunnan Province and Tibetan Plateau, respectively. Based on the estimated high genetic variability, distinct clustering in BA phylogeny, and distinct haplotypes, the genetic information suggested the distinctness of the D. gracilis population in northeast India. The present study encourages more extensive sampling of D. gracilis from different geographical locations and the generation of multiple genetic marker data or high- throughput sequence information to elucidate the actual diversity of this reptile species. The current genetic information not only evidenced the possible cryptic diversity of D. gracilis from the Indochina sub-region, but also invites to begin other biological and ecological studies to facilitate the evolutionary understanding of this species, and the holistic conservation action plans needed to protect them in nature.
Acknowledgements
We are thankful to the Director of Zoological Survey of India (ZSI), Ministry of Environment, Forest and Climate Change (MoEF&CC), the Government of India, for providing necessary facilities and support for the study. We are also thankful to the anonymous reviewer and Vishal Santra, Reptile Consultant, India and member of IUCN Viper Specialist Group for helping in improving the language.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI GenBank database at (https://www.ncbi.nlm.nih.gov) with the accession number (MT450474) which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Additional information
Funding
References
- Antão LH, Bates AE, Blowes SA, Waldock C, Supp SR, Magurran AE, Dornelas M, Schipper AM. 2020. Temperature-related biodiversity change across temperate marine and terrestrial systems. Nat Ecol Evol. 4(7):927–933.
- Brygooé R. 1987. Les Ophisaurus (Sauria, Anguidae) d’Asieorientale. Bulletin du Museum d’Histoire Naturelle, Paris. 4:727–752.
- Burbrink FT, Lawson R. 2007. How and when did old world ratsnakes disperse into the New World? Mol Phylogenet Evol. 43(1):173–189.
- Bush A, Sollmann R, Wilting A, Bohmann K, Cole B, Balzter H, Martius C, Zlinszky A, Calvignac-Spencer S, Cobbold CA, et al. 2017. Connecting Earth observation to high-throughput biodiversity data. Nat Ecol Evol. 1(7):176.
- Cai B, Guo X, Jiang J. 2020. Next-generation sequencing yields a complete mitochondrial genome of the Asian Glass Lizard (Dopasia gracilis) from the Yungui Plateau in Southwest China. Mitochondrial DNA Part B. 5(1):992–993.
- Carvalho SB, Velo-Antón G, Tarroso P, Portela AP, Barata M, Carranza S, Moritz C, Possingham HP. 2017. Spatial conservation prioritization of biodiversity spanning the evolutionary continuum. Nat Ecol Evol. 1(6):151.
- Chambers EA, Hebert PDN. 2016. Assessing DNA barcodes for species identification in North American reptiles and amphibians in natural history collections. PLOS One. 11(4):e0154363.
- Clement M, Snell Q, Walker P, Posada D, Crandall K. 2000. TCS: a computer program to estimate gene genealogies. Mol Ecol. 9(10):1657–1659.
- Conrad JL, Ast JC, Montanari S, Norell MA. 2011. A combined evidence phylogenetic analysis of Anguimorpha (Reptilia: Squamata). Cladistics. 27(3):230–248.
- Conrad JL. 2008. Phylogeny and systematics of Squamata (Reptilia) based on morphology. Bullet Am Museum Nat Hist. 310:1–182.
- Das I, Das A. 2017. A naturalist’s guide to the reptiles of India, Bangladesh, Bhutan, Nepal, Pakistan and Sri Lanka. Oxford: John Beaufoy Publishing Ltd.
- Gray JE. 1845. Catalogue of the specimens of lizards in the collection of the British Museum. London: Trustees of die British Museum/Edward Newman.
- Guo P, Liu Q, Xu Y, Jiang K, Hou M, Ding L, Pyron RA, Burbrink FT. 2012. Evolution out of Asia: natricine snakes support the Cenozoic Beringian dispersal hypothesis. Mol Phylogenet Evol. 63(3):825–883.
- Gvoždík V, Benkovský N, Crottini A, Bellati A, Moravec J, Romano A, Sacchi R, Jandzik D. 2013. An ancient lineage of slow worms, genus Anguis (Squamata: Anguidae), survived in the Italian Peninsula. Mol Phylogenet Evol. 69(3):1077–1092.
- Gvoždík V, Jandzík D, Lymberakis P, Jablonski D, Moravec J. 2010. Slow worm, Anguis fragilis (Reptilia: Anguidae) as a species complex: genetic structure reveals deep divergences. Mol Phylogenet Evol. 55(2):460–472.
- IUCN. 2020. The IUCN red list of threatened species. Version 2020.1. [accessed 2020 May 14]. https://www.iucnredlist.org
- Jablonski D, Jandzik D, Mikulíček P, Džukić G, Ljubisavljević K, Tzankov N, Jelić D, Thanou E, Moravec J, Gvoždík V. 2016. Contrasting evolutionary histories of the legless lizards slow worms (Anguis) shaped by the topography of the Balkan Peninsula. BMC Evol Biol. 16:99.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Kundu S, Kumar V, Laskar BA, Tyagi K, Chandra K. 2018. Pet and turtle: DNA barcoding identified twelve Geoemydid species in northeast India. Mitochondrial DNA Part B. 3(2):513–518.
- Lalremsanga HT, Khawlhring L, Lalrotluanga . 2010. Three additional lizard (Squamata: Sauria) records for Mizoram, India. J Threat Taxa. 2:718–720.
- Laopichienpong N, Muangmai N, Supikamolseni A, Twilprawat P, Chanhome L, Suntrarachun S, Peyachoknagul S, Srikulnath K. 2016. Assessment of snake DNA barcodes based on mitochondrial COI and Cytb genes revealed multiple putative cryptic species in Thailand. Gene. 594(2):238–247.
- Lavin BR, Girman DJ. 2019. Phylogenetic relationships and divergence dating in the Glass Lizards (Anguinae). Mol Phylogenet Evol. 133:128–140.
- Leigh JW, Bryant D. 2015. Popart: full-feature software for haplotype network construction. Methods Ecol Evol. 6(9):1110–1116.
- Letunic I, Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 23(1):127–128.
- Lin S-M, Chang W-S, Chen S-L, Shang G, Lue K-Y. 2003. Taxonomic status of the legless lizard Ophiosaurus (Squamata: Anguidae) in Taiwan: molecular data, morphology, and literature review. Zool Stud. 42:411–419.
- Macey JR, Schulte JA, Larson A, Tuniyev BS, Orlov N, Papenfuss TJ. 1999. Molecular phylogenetics, tRNA evolution, and historical biogeography in anguid lizards and related taxonomic families. Mol Phylogenet Evol. 12(3):250–272.
- McConkey EH. 1954. A systematic study of North American lizards of the genus Ophisaurus. Am Midland Nat. 51(1):133–171.
- Mulcahy DG, Noonan BP, Moss T, Townsend TM, Reeder TW, Sites JW, Wiens JJ. 2012. estimating divergence dates and evaluating dating methods using phylogenomic and mitochondrial data in squamate reptiles. Mol Phylogenet Evol. 65(3):974–991.
- Nagy ZT, Sonet G, Glaw F, Vences M. 2012. First large-scale DNA barcoding assessment of reptiles in the biodiversity hotspot of madagascar, based on newly designed COI primers. PLOS One. 7(3):e34506.
- Nguyen TQ, Bohme W, Nguyen TT, Le QK, Pahl KR, Haus T, Ziegler T. 2011. Review of the genus Dopasia Gray, 1853 (Squamata: Anguidae) in the Indochina subregion. Zootaxa. 2894(1):58–68.
- Nguyen VS, Ho TC, Nguyen QT. 2009. Herpetofauna of Vietnam. Frankfurt am Main: Edition Chimaira.
- Nylander JAA. 2004. Mr.Modeltest v2, Program distributed by the author. Uppsala: Evolutionary Biology Centre, Uppsala University.
- Pan HC, Liu L, Li P, Li XF, Liu ZL. 2015. The complete mitochondrial genome of Chinese glass lizard Ophisaurus harti (Squamata: Anguidae)). Mitochondrial DNA. 26(2):280–281.
- Pough FH, Andrews RM, Cadle JE, Crump ML, Savitzky AH, Wells KD. 1998. Herpetology. Upper Saddle River, NJ: Prentice-Hall.
- Pyron RA, Burbrink FT, Wiens JJ. 2013. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol Biol. 13:93
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Rozas J, Ferrer-Mata A, Sanchez-DelBarrio J, Guirao-Rico S, Librado P, Ramos-Onsins S, Sanchez-Gracia A. 2017. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 34(12):3299–3302.
- Sanmartín I, Enghoff H, Ronquist F. 2001. Patterns of animal dispersal, vicariance and diversification in the Holarctic. Biol J Linn Soc. 73(4):345–390.
- Song B, Cheng S, Sun Y, Zhong X, Jin J, Guan R, Murphy RW, Che J, Zhang Y, Liu X. 2015. A genome draft of the legless anguid lizard, Ophisaurus gracilis. Gigascience. 4:17
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25(24):4876–4882.
- Townsend TM, Alegre RE, Kelley ST, Wiens JJ, Reeder TW. 2008. Rapid development of multiple nuclear loci for phylogenetic analysis using genomic resources: an example from squamate reptiles. Mol Phylogenet Evol. 47(1):129–142.
- Uetz P, Freed P, Hošek J. 2020. The reptile database. [accessed 2020 May 14]. http://www.reptile-database.org.
- Vidal N, Hedges SB. 2005. The phylogeny of squamate reptiles (lizards, snakes, and amphisbaenians) inferred from nine nuclear protein-coding genes. CR Biologies. 328(10–11):1000–1008.
- Vidal N, Hedges SB. 2009. The molecular evolutionary tree of lizards, snakes, and amphisbaenians. CR Biologies. 332(2–3):129–139.
- Wiens JJ, Kuczynski CA, Townsend T, Reeder TW, Mulcahy DG, Sites JW. 2010. Combining Phylogenomics and fossils in higher-level squamate reptile phylogeny: molecular data change the placement of fossil taxa. Syst Biol. 59(6):674–688.
- Wiens JJ, Slingluff JL. 2001. How lizards turn into snakes: a phylogenetic analysis of body-form evolution in anguid lizards. Evolution. 55(11):2303–2318.
- Wüster W, Peppin L, Pook CE, Walker DE. 2008. A nesting of vipers: phylogeny and historical biogeography of the Viperidae (Squamata: Serpentes). Mol Phylogenet Evol. 49(2):445–459.
