Abstract
Sorbus insignis (Hook. f.) Hedl., belonging to S. sect. Multijugae Yu is an epiphytic shrub. Its phylogenetic position is still poorly understood. In this study, we assembled its complete chloroplast genome from whole-genome high-throughput sequencing data. The chloroplast genome was 159, 993 bp in length, with a large single-copy (LSC) region of 87, 932 bp, a small single-copy (SSC) region of 19, 255 bp, separated by two inverted repeat (IR) regions of 26, 403 bp each. It was predicted to contain a total of 132 genes, with an overall GC content of 36.56%. Phylogenetic analysis suggested S. insignis belongs to S. L. sensu stricto and closest to S. prattii Koehne among the published chloroplast genome.
Sorbus insignis (Hook. f.) Hedl., belonging to S. sect. Multijugae Yu (Rosaceae) is a shrub native to the southwest of China, north of Myanmar, north of India, and Nepal (Yu and Lu Citation1997; Lu et al. Citation2003). This species is quite distinct in genus Sorbus, which occurs epiphytic habits usually climbing along the trunk and branch of broadleaf trees or cliff and rocks, and distributed at an altitude from 2600 to 3300 m. The morphological and systematics studies indicated S. insignis is most closely related to S. Harrowiana (Balf. F. et W. W. Smith) Rehd., a species character in larger leaf and distributed along Gaoligong Mountain (Yu et al. Citation1997; Lu and Stephen Citation2003; Lo and Donoghue Citation2012; Li et al. Citation2017). While, some flora and taxonomy study treated the S. harrowiana as a synonym of S. insignis (Lu and Stephen Citation2003). This taxonomic confusion could be solved by phylogenetic studies. In this study, the complete chloroplast genome sequence of S. insignis was sequenced and characterized. We also constructed a phylogenetic tree to confirm its relationship with other species within the genus Sorbus. The annotated genome sequence is accessible from GenBank with the accession number (GenBank: MT677871).
Fresh leaves of S. insignis were sampled in Gaoligong Mountain National Nature Reserve, Lushui city, Yunnan Province, China (25.980275 N, 98.696044E). The voucher specimen (B.Y. Liao & W.Y. Zhao 2018084) was deposited in the Herbarium of Sun Yat-sen University (SYS). Total genomic DNA was extracted from silica-dried leaves using the modified CTAB method. The DNA library was prepared with a TruSeq DNA Sample Prep Kit (Illumina, USA) according to the instructions of the manufacturer. Then the DNA library was sequenced on an Illunima Hiseq 2500 Sequencing System (Guangzhou, China). A total of 6 Gb paired-end sequence (150 bp) was generated and used to assemble the chloroplast genome in GetOrganelle (Jin et al. Citation2018). The genome annotation was performed by the Geseq online tool (Tillich et al. Citation2017) and Geneious ver. 10.1 (Kearse et al. Citation2012), then manually verified and corrected by comparison with sequences of related species, for example, S. tianschanica (GenBank: MK920289), S. prattii (GenBank: MK814479), and S. tianschanica (GenBank: MK920289).
The circular chloroplast genome S. insignis was 159,993 bp in length, with a large single-copy (LSC) region of 87, 932 bp, a small single-copy (SSC) region of 19, 255 bp, separated by two inverted repeat (IR) regions of 26, 403 bp each. It was predicted to contain 132 genes, including 87 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The overall GC content was 36.56%.
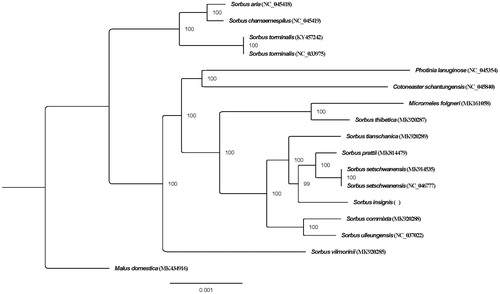
To investigate the relationship of S. insignis, the chloroplast genomes of S. insignis and 16 other species were aligned using MAFFT ver. 7.307 (Katoh and Standley Citation2013), and the Malus domestica was selected as outgroup. A phylogenetic tree () was constructed with the maximum likelihood method using RAxML (Stamatakis Citation2014). The result of the phylogenetic analysis revealed that Sorbus L. sensu lato is not monophyletic (), which is consistent with previous research results (Lo and Donoghue Citation2012; Li et al. Citation2017). The S. insignis is nested within S. L. sensu stricto and sister to S. prattii Koehne and S. setschwanensis (Schneid.) Koehne. S. insignis were previously placed in S. Ser. Insignes Yu (Yu and Kuan Citation1963), while a molecular systematics bases on four nuclear and one chloroplast marker suggest to place it into S. Ser. Multijugae Yu (Li et al. Citation2017). Our results support S. insignis belongs to S. L. sensu stricto. The chloroplast genome of S. insignis reported here provides new resources for the phylogenetic study of the genus Sorbus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT677871.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. bioRxiv. 256479.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li M, Ohi-Toma T, Gao YD, Xu B, Zhu ZM, Ju WB, Gao XF. 2017. Molecular phylogenetics and historical biogeography of Sorbus sensu stricto (Rosaceae). Mol Phylogenet Evol. 111:76–86.
- Lo EYY, Donoghue MJ. 2012. Expanded phylogenetic and dating analyses of the apples and their relatives (Pyreae, Rosaceae). Mol Phylogenet Evol. 63(2):230–243.
- Lu LD, Stephen AS. 2003. Flora of China. Volume 9. St. Louis, MO, USA: Missouri Botanical Garden Press. Beijing, China: Science Press.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Yu DJ, Lu LD. 1997. Rosaceae. In: Chen C, Chang H, Miau R, Hsu T, editors. Flora Reipublicae Popularis Sinicae. Beijing, China: Science Press. Volume 43, p. 312–313.
- Yu TT, Kuan KJ. 1963. Taxa nova Rosacearum sinicarum (I). Acta Phytotaxonomica Sinica. 8(3):202–234.