Abstract
Phyllostachys glauca is a dominant species in limestone mountains endemic to China. Here, we characterized its complete chloroplast genome. It is a circular DNA molecule of 139,689 bp in length, including a pair of 21,798 bp inverted repeats (IRs), a 12,872 bp small single-copy (SSC) region and an 83221 bp large single-copy (LSC) region. The total GC content of P. glauca chloroplast genome was 38.9%, and it encodes a total of 137 functional genes, including 89 protein-coding genes, 40 tRNA genes, and 8 rRNA genes. The phylogenetic analysis shows that P. glauca is highly clustered in the Phyllostachys clade (V), sister to the lineage of P. nigra var. henonis + P. sulphurea.
Limestone area is usually ecologically fragile complex terrains, where the topography considerably affects vegetation coverage (Pu et al. Citation2017). Adapted to drought stress by regulating the concentration and distribution of nutrient elements, Phyllostachys glauca, a temperate woody bamboo, is one of the dominant species in limestone mountains of southern China (Du et al. Citation1994; Liang et al. Citation2019). Like most bamboo plants, P. glauca shows a pattern of massive synchronized flowering and leads to large-scale death after flowering (Zheng et al. Citation2020).
In this study, we obtained and reported the complete chloroplast genome of P. glauca. Fresh and young leaves as sequencing materials were collected from the bamboo garden of Jiangxi Agricultural University, China (28°45′40″N, 115°49′31″E). The voucher specimen (JXAU-20201532) was deposited at the herbarium of the College of Forestry, Jiangxi Agricultural University, China. Illumina paired-end (PE) library was prepared and sequenced in the Nanjing Novogene Bio-technology Co., Ltd., China. The clean reads were assembled by using GetOrganelle v1.5 (Jin et al. Citation2018). Subsequently, the plastome was annotated using GeSeq (Tillich et al. Citation2017) and Geneious 9.0.5 (http://www.geneious.com/), and detected simple sequence repeats (SSR) by MISA (http://pgrc.ipk-gatersleben.de/misa). The annotated complete chloroplast genome of P. glauca was deposited in GenBank under the accession no. MT657329.
The chloroplast genome of P. glauca is a typical circular structure of 139,689 bp in length, and consisting of a large single-copy region with 83,221 bp (LSC), a small single-copy region with 12,872 bp (SSC), and two inverted repeat regions with 21,798 bp (IRs). The total GC content of the chloroplast genome is 38.9%, meanwhile the corresponding value of LSC, SSC, and IR regions is 37.0%, 33.2% and 44.2%, respectively. The whole genome contained 137 functional genes, including 89 protein-coding genes, 40 tRNA genes, and 8 rRNA genes.
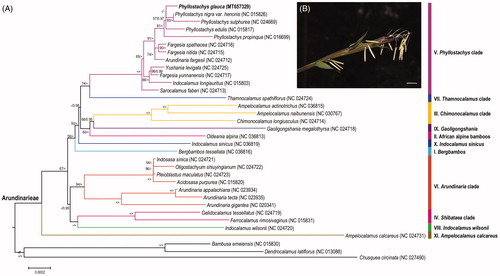
To evaluate the phylogenetic position of P. glauca, additional 30 complete chloroplast genomes in the trib. Arundinarieae, together with three species as outgroup, were retrieved from NCBI (). All sequences were aligned with MAFFT 7.409 (Katoh and Toh Citation2010). Maximum-likelihood tree and Bayes tree were constructed using RAxML 8.2.8 (Stamatakis Citation2014) and MrBayes 3.2.6 (Ronquist and Huelsenbeck Citation2003), respectively. The results showed that P. glauca is highly clustered in the Phyllostachys clade (V), as a sister group with the lineage of P. nigra var. henonis and P. sulphurea. In addition, the relationships within Phyllostachys genus are still weakly supported in the phylogenetic tree with short internodes, reflecting the probable recent rapid radiation during the evolution.
Figure 1. Phylogenetic position and flowers of P. glauca. A. Maximum likelihood tree based on protein coding sequences of chloroplast genomes from 34 bamboos. Numbers associated with branches are ML bootstrap values, and Bayesian posterior probabilities, respectively. ‘*’ indicate 100% bootstrap support or 1.0 posterior probability. ‘-’ indicate the bootstrap support or posterior probability lower than 50% or 0.5. B. Flowering branches of P. glauca, showing mixed inflorescence. Scale bar = 10 mm.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available in ‘Dryad’ at http://doi:10.5061/dryad.m37pvmd07, reference number [m37pvmd07].
Additional information
Funding
References
- Du TZ, Li ZY, Yang GY, Zhu WH, Liu XW, Cheng DY, Fang L. 1994. Study on site of Phyllostachys glauca in limestone area. Acta Agric Univ Jiangxiensis. 16(1):82–87.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 4:256479.
- Katoh K, Toh H. 2010. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 26(15):1899–1900.
- Liang K, Fan Y, Feng HJ, Tan TT, Shi JM. 2019. Concentration and distribution pattern of non-structural carbohydrate of Phyllostachys glauca in different limestone habitats. Scientia Silvae Sinicae. 55(6):22–27.
- Pu G, Lv Y, Xu G, Zeng D, Huang Y. 2017. Research progress on Karst tiankeng ecosystems. Bot Rev. 83(1):5–37.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Zheng X, Lin S, Fu H, Wan YW, Ding YL. 2020. The bamboo flowering cycle sheds light on flowering diversity. Front Plant Sci. 11(381).
