Abstract
The paper wasp Polistes riparius is distributed in cold regions of northern East Asia to Russia. P. riparius are characterized by having longer cells than those of the closely related P. chinensis, which has a similar life history, as an adaptation to cold regions. The phylogenetic relationships of paper wasps have recently been studied; however, the genetic diversity and population structure of P. riparius has not been determined. The present study is the first to analyze the complete mitochondrial genome using next generation sequencing of P. riparius collected from Sapporo, Hokkaido Prefecture, Japan. The genome consisted of a closed loop that was 16,383 bp-long and included 13 protein coding genes (PCGs), 22 tRNA genes, two rRNA genes, and one AT-rich control region. The average AT content was 84.54%. The heavy (H)-strand was predicted to have 12 PCGs and 14 tRNA genes, while the light (L)-strand was predicted to contain one PCGs, eight tRNA genes, and two rRNA genes. All PCGs started with ATG. Stop codons were of two types: TAA for 11 genes (ND1, ND2, ND3, ND4L, ND5, ND6, COXI, COXII, COXIII, COB, ATP6 and ATP8) and TAG for two genes (ND3 and ND4). The molecular phylogenetic relationship based on the maximum likelihood method using 13 PCGs was consistent with some previous studies in which a closely relationship between P. riparius and P. jokahamae.
Polistes is a species-rich genus of paper wasps that includes more than 200 species (Carpenter Citation1996). They have long been treated as model materials for studying cooperation, competitive breeding, and social evolution in Hymenoptera because they are relatively nonaggressive and have small observable colonies with exposed nests. Polistes riparius is distributed in the cold regions of northern East Asia to Russia (Yamane and Yamane Citation1987; Kurzenko Citation1995; Dubatolov Citation1998). Although P. riparius has a life history that is similar to the closely related P. chinensis, it is characterized by developing longer cells than those of P. chinensis, possibly as a heat-retaining mechanism (Yamane and Kawamichi Citation1975; Yamane et al. Citation1998). The phylogenetic relationships of paper wasps were recently studied based on the analysis of partial sequences of mitochondrial DNA (Santos et al. Citation2015). Currently the genetic diversity and population structure of this species, and its characteristic ecological adaptations to cold regions, have not been identified. Our study analyzed the complete mitochondrial genome of P. riparius.
We collected adult P. riparius workers on flowers in Sapporo, Hokkaido Prefecture, Japan, in 2013 (42°57′N, 141°15′E). Adult workers were transferred immediately to 99.5% ethanol for mitochondrial DNA analysis. The specimens were stored in a freezer at −20 °C in our laboratory at the Kyoto Sangyo University. Genomic DNA isolated from one worker was sequenced using the Illumina MiSeq platform (Illumina, San Diego, CA, USA). The complete mitochondrial genome of the paper wasp P. jokahamae was used as a reference sequence. The resultant reads were assembled and annotated using the Geneious R9 software (Kearse et al. Citation2012) and the MITOS web server (Bernt et al. Citation2013). Thirteen protein-coding genes (PCGs) and two rRNA genes sequences were aligned using Genetyx version 15 (GENEYTX, Japan). The phylogenetic analysis was based on the nucleotide sequences of 13 protein-coding genes using MEGAX (Kumar et al. Citation2018).
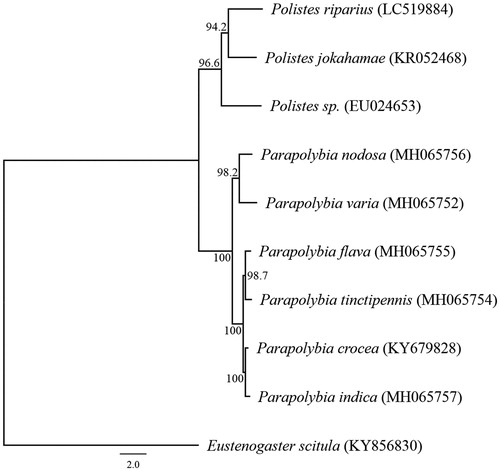
We succeeded in sequencing the entire mitochondrial genome of P. riparius from Hokkaido Prefecture (DDBJ accession number LC519884). These specimens were stored as stocks at the National Museum of Nature and Science, Japan (control number NSMT-I-Hym75321). The genome consisted of a closed loop 16,383 bp long and included 13 PCGs, 22 tRNA genes, two rRNA genes, and one AT-rich control region that represented a typical Vespidae mitochondrial genome. The average AT content was 84.54%. The heavy strand was predicted to have 12 PCGs and 14 tRNA, while the light strand was predicted to contain one PCG, eight tRNA genes, and two rRNA genes. The start codon was ATG for the 13 PCGs. The stop codon was TAA for 11 PCGs, and TAG for two genes. The genes ATP8 and ATP6 shared 10 nucleotides, and ATP6 and COIII shared one nucleotide. The genes ND4 and ND4L, and genes ND5 and ND6 shared 7 and 4 nucleotides, respectively. A molecular phylogenetic analysis of 9 closely related taxa of Vespidae suggested sister relationships within the Polistes (). The complete sequence of the mitochondrial DNA of P. riparius provides additional genetic information for studying the phylogenetic relationship of the genus Polistes.
Figure 1. Phylogenetic relationships (maximum likelihood) of the Vespidae based on the nucleotide sequences of the 13 protein-coding genes of the mitochondrial genome. Sequences from Eustenogaster scitula (KY856830) was used as an outgroup. These sequences were separated by codon positions, and for each partition, the optimal models of sequence evolution were used in the maximum likelihood method using MEGA X, based on the corrected Akaike information criterion. The numbers at the nodes indicate the bootstrap support inferred from 1000 bootstrap replicates. Alphanumeric terms indicate the DNA Database of Japan accession numbers.
Acknowledgements
We are grateful to Prof. Koji Tsuchida of Gifu University for helping our research.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Data availability statement
The data that support the findings of this study are openly available in DDBJ/GenBank at https://www.ddbj.nig.ac.jp/index.html, accession number LC519884.
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Carpenter JM. 1996. Distributional checklist of species of the genus Polistes (Hymenoptera: Vespidae; Polistinae, Polistini). Am Mus Nov. 3188:1–39.
- Dubatolov VV. 1998. Social wasps (Hymenoptera, Vespidae: Polistinae, Vespinae) of Siberia in the collection of Siberian Zoological Museum. Far Eastern Entomologist. 57:1–11.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Kurzenko NV. 1995. Fam. Vespidae. In: A key to insects from the Far East of Russia. Vol. 4. Neuropteroidea, Mecoptera, Hymenoptera. Part 1. Nauka: St-Petersburg :264–324. (In Russian).
- Santos BF, Payne A, Pickett KM, Carpenter JM. 2015. Phylogeny and historical biogeography of the paper wasp genus Polistes (Hymenoptera: Vespidae): implications for the overwintering hypothesis of social evolution. Cladistics. 31(5):535–549.
- Yamane S, Yamane S. 1987. A new species and new synonymy in the subgenus Polistes of eastern Asia (Hymenoptera, Vespidae). Kontyû. 55:215–219.
- Yamane S, Kawamichi T. 1975. Bionomic comparison of Polistes biglumis (Hymenoptera, Vespidae) at two different localities in Hokkaido, northern Japan, with reference to its probable adaptation to cold climate. Kontyû. 43:214–232.
- Yamane S, Kudô K, Tajima T, Nihon’yanagi K, Shinoda M, Saito K, Yamamoto H. 1998. Comparison of investment in nest construction by the foundresses of consubgeneric Polistes wasps, P. (Polistes) riparius and P. (P.) chinensis (Hymenoptera, Vespidae). J Ethol. 16(2):97–104.
