Abstract
Haemaphysalis longicornis (Ixodida: Ixodidae) is a common blood-feeding ectoparasite of the giant panda and poses significant health burden to wild and captive populations. In the present study, the complete mitogenome of the giant panda tick H. longicornis was sequenced using Illumina sequencing technology. The entire mitogenome was 14,706 bp in length and encoded 37 genes including 13 protein-coding genes, 22 transfer RNAs and two ribosomal RNAs. Phylogeny showed that four isolates of H. longicornis, regardless of host origins or locations, clustered together and had a closer relationship with Haemaphysalis hystricis than other Haemaphysalis species among the subfamily Haemaphysalinae of Ixodidae. The cumulative mitochondrial DNA resources provide insights into genetic and phylogenetic studies of Haemaphysalis ticks.
The giant panda (Ailuropoda melanoleuca) is a flagship species for wildlife conservation in the world (O’Brien et al. Citation1994; Wang et al. Citation2018). The ticks are common blood-feeding ectoparasites found in giant pandas and poses significant health burden to wild and captive populations due to dermatitis, anemia and other blood-borne infectious diseases caused by their vector roles in nature (de la Fuente et al. Citation2008; Cheng et al. Citation2013; Wang et al. Citation2018). Until now more than 13 tick species have been identified from giant pandas based on morphological characteristics (Cheng et al. Citation2013; Wang et al. Citation2018; Liu et al. Citation2020). Although morphological identification seems convenient and valid, it is labor-consuming, time-costing and often be unrecognized even by experienced microscopists, especially in assessing the close-related tick species that infest wild animals (Qin et al. Citation2011). Molecular approach using mitochondrial (mt) DNA is proven to be a valuable complementary tool for overcoming this limitation and has been recently used for identification and characterization of ticks (Hwang et al. Citation2001; Tian et al. Citation2011; Cheng et al. Citation2013; Liu et al. Citation2013; Burger et al. Citation2014; Liu et al. Citation2020). In the present study, we reported the complete mitogenome sequence of the giant panda tick Haemaphysalis longicornis and aimed to provide novel mitochondrial source to this ectoparasite.
On March 25, 2020, five tick specimens were collected from a captive and recently rescued giant panda in the Dujiangyan Base of the China Conservation and Research Center for the Giant Panda, Sichuan Province of Southwest China (30°59′N, 103°37′E). The ticks (one male and four females) were identified as H. longicornis according to morphological keys of Tanskul and Inlao (Citation1989) and molecular sequencing of the mt 16S and ribosomal ITS2 genes (Tian et al. Citation2011). One female tick was used for DNA extraction and the others were archived in the Parasitological Museum of Sichuan Agricultural University (Sichuan, China) under collection numbers XY2018_15-18. The mitogenome was sequenced by the Illumina HiSeq2500 platform (Novogene, Tianjin, China), assembled using MITObim (Hahn et al. Citation2013) and annotated by MITOS (Bernt et al. Citation2013). The complete genome sequence was deposited in GenBank under accession number: MT780294.
The complete mitogenome of H. longicornis was 14,706 bp in length with 77.2% AT and encoded 13 protein-coding genes (PCGs), 22 tRNA genes and two rRNA genes. Among the 37 genes, four PCGs, two rRNAs and eight tRNAs were located on the forward strand (H-strand), while the remaining genes were transcribed on the reverse strand (L-strand). Thirteen PCGs, except nad6 deduced to use an incomplete stop codon ‘T’, were predicted to use the typical TAA or TAG as the stop codons. Twenty-two tRNA genes ranged from 53 bp (tRNA-Cys) to 74 bp (tRNA-Met) in length and had typical clover-leaf like secondary structures. Two rRNA genes were 659 bp (12S) and 1,187 bp (16S) in lengths, respectively, and placed between tRNA-Leu and tRNA-Ile with a separation by tRNA-Val. The control region (also known as D-loop region) was located between tRNA-Leu and tRNA-Cys, similar to other Haemaphysalis ticks, suggesting its conservation and function in regulation of transcription and control of DNA replication (Clayton Citation1991).
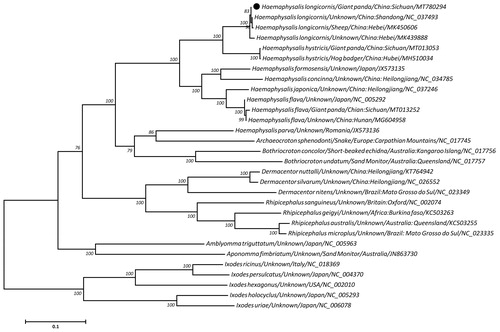
Building on a concatenated amino acid sequences of 13 protein-coding genes from H. longicornis and 29 other ticks, the maximum-likelihood (ML)-based phylogeny demonstrated that four isolates of H. longicornis, regardless of host origins or locations, clustered together and shared a more close relationship to Haemaphysalis hystricis than to other Haemaphysalis ticks, with 100% bootstrap confidence (), supporting their species validity in the subfamily Haemaphysalinae. In addition, the genera including Archaeocroton, Bothriocroton, Dermacentor, Rhipicephalus, Amblyomma and Aponomma were treated as monophyletic relationships with Haemaphysalis in the family Ixodidae, agreement with recent molecular studies (Burger et al. Citation2013; Geng et al. Citation2017; Tian et al. Citation2019; Liu et al. Citation2020). In summary, the H. longicornis mitogenome sequenced here provides novel insights into genetic and phylogenetic studies of this tick.
Figure 1. Maximum likelihood tree inferred from the concatenated amino-acid sequences of 13 mitochondrial PCGs of H. longicornis and other related ticks, utilizing MtArt + G + I model and 1,000,000 bootstrap replications. Ixodes species were used as outgroups. Values lower than 50% are not shown. The black cycle represents the species in this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT780294.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Burger TD, Shao R, Barker SC. 2013. Phylogenetic analysis of the mitochondrial genomes and nuclear rRNA genes of ticks reveals a deep phylogenetic structure within the genus Haemaphysalis and further elucidates the polyphyly of the genus Amblyomma with respect to Amblyomma sphenodonti and Amblyomma elaphense. Ticks Tick Borne Dis. 4(4):265–274.
- Burger TD, Shao R, Barker SC. 2014. Phylogenetic analysis of mitochondrial genome sequences indicates that the cattle tick, Rhipicephalus (Boophilus) microplus, contains a cryptic species. Mol Phylogenet Evol. 76:241–253.
- Cheng WY, Zhao GH, Jia YQ, Bian QQ, Du SZ, Fang YQ, Qi MZ, Yu SK. 2013. Characterization of Haemaphysalis flava (Acari: Ixodidae) from Qingling subspecies of giant panda (Ailuropoda melanoleuca qinlingensis) in Qinling mountains (Central China) by morphology and molecular markers. PLoS One. 8(7):e69793.
- Clayton DA. 1991. Replication and transcription of vertebrate mitochondrial DNA. Annu Rev Cell Biol. 7:453–478.
- de la Fuente J, Estrada-Pena A, Venzal JM, Kocan KM, Sonenshine DE. 2008. Overview: ticks as vectors of pathogens that cause disease in humans and animals. Front Biosci. 13:6938–6946.
- Geng J, Zheng A, Zou Z, Zhang X. 2017. The complete mitochondrial genome and phylogenetic analysis of Haemaphysalis longicornis Neumann (Acari: Ixodidae). Mitochondrial DNA B Resour. 2(2):856–857.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Hwang UW, Park CJ, Yong TS, Kim W. 2001. One-step PCR amplification of complete arthropod mitochondrial genomes. Mol Phylogenet Evol. 19(3):345–352.
- Liu GH, Chen F, Chen YZ, Song HQ, Lin RQ, Zhou DH, Zhu XQ. 2013. Complete mitochondrial genome sequence data provides genetic evidence that the brown dog tick Rhipicephalus sanguineus (Acari: Ixodidae) represents a species complex. Int J Biol Sci. 9(4):361–369.
- Liu Y, Wang L, Wang L, Deng L, Wei M, Wu K, Huang S, Li G, Huang Y, Zhang H, et al. 2020. Characterization of the complete mitogenome sequence of the giant panda tick Haemaphysalis hystricis. Mitochondrial DNA Part B Resour. 5(2):1191–1193.
- O’Brien SJ, Pan W, Lu Z. 1994. Pandas, people and policy. Nature. 369:179–180.
- Qin XC, Tian JH, Wang JB, Lu X, Sun QZ, Jin D, Zhou DJ, Xu JG, Zhang YZ. 2011. Identification of Haemaphysalis longicornis and Rhipicephalus microplus. Chin J Epidemiol. 32:608–612.
- Tanskul P, Inlao I. 1989. Keys to the adult ticks of Haemaphysalis Koch, 1844, in Thailand with notes on changes in taxonomy (Acari: Ixodoidea: Ixodidae). J. Med. Entomol. 26(6):573–600.
- Tian J, Ge M, Xu H, Wu T, Yu B, Lei C. 2019. The complete mitochondrial genome and phylogenetic analysis of Haemaphysalis hystricis (Parasitiformes: Ixodidae). Mitochondrial DNA B Resour. 4(1):1049–1050.
- Tian Z, Liu G, Xie J, Yin H, Luo J, Zhang L, Zhang P, Luo J. 2011. Discrimination between Haemaphysalis longicornis and H. qinghaiensis based on the partial 16S rDNA and the second internal transcribed spacer (ITS-2). Exp Appl Acarol. 54(2):165–172.
- Wang T, Xie Y, Zheng Y, Wang C, Li D, Koehler AV, Gasser RB. 2018. Parasites of the giant panda: a risk factor in the conservation of a species. Adv Parasitol. 99:1–33.
