Abstract
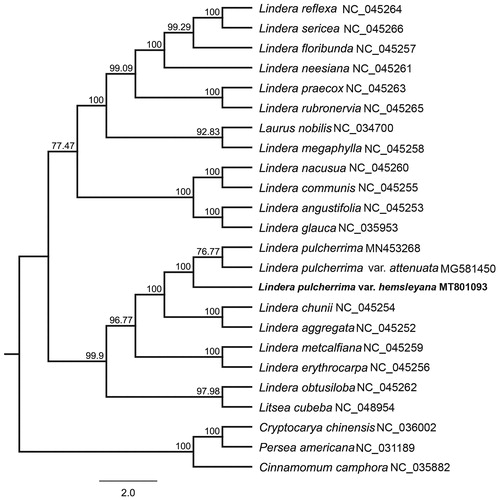
Lindera pulcherrima var. hemsleyana is an evergreen tree of the genus Lindera with important medicinal value. The complete chloroplast genome of L. pulcherrima var. hemsleyana is 152,710 bp in length, containing a LSC region of 93,751 bp, a SSC region of 18,813 bp, and a pair of inverted repeats (IRA and IRB) of 20,073 bp each. The genome encoded 128 genes, including 83 protein-coding genes, 8 ribosomal RNA genes, 36 transfer RNA genes, and one pseudogene (ycf1). Most of these genes occurred in a single copy, whereas 15 genes occurred in double copies, including all rRNA, 6 tRNA, and 5 protein-coding genes. The GC content of the whole genome, LSC, SSC, and IR regions is 39.2%, 38.0%, 33.9%, and 44.4%, respectively. A total of 90 SSRs were discovered, the numbers of mono-, di-, tri-, tetra-, penta-, and hexa-nucleotides SSRs were 66, 10, 4, 8, 1, and 1, respectively. Phylogenetic analysis of cp genomes from 24 species of Lauraceae revealed that cp genomes of L. pulcherrima var. hemsleyana, L. pulcherrima var. attenuata, and L. pulcherrima formed a monophyletic clade with 100% bootstrap value.
Lindera pulcherrima (Nees) J. D. Hooker (Lauraceae), a dominant evergreen tree in shady forests, is an important medicinal plant distributed in southern China, Nepal, and India. The active compounds contained in L. pulcherrima have good anti-inflammatory, antiviral, antibacterial, and antitumor pharmacological activities (Joshi and Mathela Citation2012). L. pulcherrima var. hemsleyana is distinguished with other two varieties, L. pulcherrima var. attenuate and L. pulcherrima var. pulcherrima, for its elliptic leaf blade, glabrous ovary, and glabrous young fruits (Tsui et al. Citation1982). The leaves and bark are used as a spice in cold, fever, and cough (Joshi and Mathela Citation2012). Its quality wood is widely used in furniture and architecture. Though phylogenic studies on Lauraceae were conducted, some taxonomic controversies on intra-generic relationship of the genus Lindera as well as the inter-generic relationship with allied genera were still unsolved (Tian et al. Citation2019).
The whole chloroplast genome was extracted from the fresh leaves of a healthy L. pulcherrima var. hemsleyana tree with modified CTAB protocol (Yang et al. Citation2014), it was collected from the natural habitats of Yongshan County (28°05′13.20″N, 103°35′38.40″E), Yunnan province, China. The voucher specimen (ELK-254) was deposited in the Herbarium of Yunnan Normal University (YNUB). Paired-end reads were sequenced by using Illumina HiSeq 2500-PE150 (Illumina, San Diego, CA). All raw reads were filtered through NGS QC toolkit_v2.3.3 to get clean reads with default parameters (Patel and Jain Citation2012). The NOVOPlasty was used to de novo assemble the clean reads to form the chloroplast genome (Dierckxsens et al. Citation2017). The assembled structures and genes of the complete cp genome were annotated with Geneious 20.0.3 (Kearse et al. Citation2012). The online tRNAscan-SE service was used to verify tRNA genes of the L. pulcherrima var. hemsleyana (Schattner et al. Citation2005).
The chloroplast genome of L. pulcherrima var. hemsleyana (GenBank accession no.: MT801093) was a quadripartite circular with 152,710 bp in size with total GC content 39.2%. A pair of inverted repeat (IR) region (20,073 bp) separated the large single copy (LSC) region (93,751 bp) and small single copy (SSC) region (18,813 bp). The GC content of the LSC, SSC, and IR regions is 38.0%, 33.9%, and 44.4%, respectively. There are 128 genes predicted, including 81 protein-coding genes, 36 tRNA genes, eight rRNA genes, and one pseudogene (ycf1). IMEx (Mudunuri and Nagarajaram Citation2007) was used to identify the SSRs with minimum repeat number set to 10, 5, 4, 3, 3, and 3. A total of 90 SSRs were discovered, the numbers of mono-, di-, tri-, tetra-, penta-, and hexa-nucleotides nucleotides SSRs were 66, 10, 4, 8, 1, and 1, respectively.
In order to understand the phylogenetic relationship between L. pulcherrima var. hemsleyana and related taxa of the genus Lindera, 19 taxa of Lindera and five species of Subfamily Lauroideae that are downloaded from NCBI as outgroups were used to construct a molecular phylogenetic tree. Twenty-four cp genomes were aligned by MAFFT 7.308 (Katoh and Standley Citation2013). The maximum likelihood (ML) tree was constructed with RAxML 8.2.11 (Stamatakis Citation2014) under the nucleotide substitution model of GTR + G. The bootstrap values were calculated from 1000 replicates analysis. The phylogenetic topology () of L. pulcherrima is consistent with the morphological delimitation of this species. L. pulcherrima var. hemsleyana and other two varieties formed one monophyletic clade with 100% bootstrap value. The complete chloroplast genome of L. pulcherrima var. hemsleyana will provide useful genetic resource for further study on phylogeny of the genus Lindera and Lauraceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/, reference number [MT801093], or available from the corresponding author.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Joshi SC, Mathela CS. 2012. Antioxidant and antibacterial activities of the leaf essential oil and its constituents furanodienone and curzerenone from Lindera pulcherrima (Nees.) Benth. ex hook. f. Pharmacogn Res. 4(2):80–84.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Mudunuri S-B, Nagarajaram H-A. 2007. IMEx: imperfect microsatellite extractor. Bioinformatics. 23(10):1181–1187.
- Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7(2):e30619.
- Schattner P, Brooks Angela N, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33(Web Server issue):W686–W689.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tian X, Ye J, Song Y. 2019. Plastome sequences help to improve the systematic position of trinerved Lindera species in the family Lauraceae. PeerJ. 7(2):e7662.
- Tsui H-P, Shia Z-D, J-L L. 1982. Lindera pulcherrima (Wall.) Benth. var. hemsleyana (Diels) H. P. Tsui in Act. Phytotax Sin. 31(4):427.
- Yang J-B, Li D-Z, Li H-T. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14(5):1024–1031.