Abstract
Artemisia scoparia Waldst. et Kitam. is an important medicinal plant of Asteraceae. Inconsistent pharmacological activities have been reported for A. scoparia from China and South Asia. As the first step to understand the underlying genetic basis, we sequences the chloroplast genome for A. scoparia collected from China and compared with one collected from Pakistan. The circular chloroplast genome of A. scoparia is 151,008 bp long, which is slightly shorter than that of A. scoparia-Pakistan. It encodes 87 protein-coding genes, 8 ribosomal RNA genes, and 37 transfer RNAs. The overall GC content of the genome is 37.47%. Comparing with A. scoparia from China and Pakistan identified the inversion of the SSC region. Besides, a total of 79 single nucleotide polymorphisms and 39 insertions and deletions were identified. The results can be used to develop molecular markers to distinguish A. scoparia from different geographical areas that might have variable bioactivities.
Artemisia scoparia Waldst. et Kitam. is an important medicinal plant of Asteraceae. It has a wild distribution in Southwest Asia and central Europe (Abad et al. Citation2012). Previous reports have shown that this species has a variety of pharmacological functions, such as anti-cancer, anti-inflammatory, anti-HBV, anti-allergic, anti-oxidant and etc (Geng et al. Citation2015; Sajid et al. Citation2017; Nam et al. Citation2018; Boudreau et al. Citation2018). In South Asia, it has been widely used in folk medicine. In contrast, A. scoparia in East Asia has mostly been used as a substitute for Artemisia annua. Whether or not the A. scoparia in China have any pharmocological activities similar to those in Southwest Asia is an interesting question. As the first step to answer the question, we sequenced the chloroplast (cp) genome of A. scoparia collected from China and compared it with those from the South Aisa (Iram et al. Citation2019).
The fresh leaves of A. scoparia were collected from Dafutuo Village, Yanqing District, Beijing, China (40°38 '77″, 151°96′ 18″ E). The specimen and its DNA were deposited at the Herbarium of Institute of Medicinal Plant Development in Beijing, China, Accession number: Implad201910001. We use the genomic DNA kit (Tiangen Biotech, Beijing) to extract the total DNA; The purity and integrity of DNA were determined by agarose gel electrophoresis. The DNA library was constructed with 1ug DNA using the library preparation kit (New England BioLabs, America), and sequenced using the Illumina Hiseq 2500 platform (Illumina, San Diego, CA). The de novo genome assembly from the clean data was accomplished utilizing the NOVOPlasty (v.2.7.2) (Dierckxsens et al. Citation2016). The correctness of the assembly was validated by mapping all clean reads to the assembly using Bowtie2 (v.2.0.1) (Langmead et al. Citation2009) under the default settings. The annotation of the chloroplast genome was conducted by using CPGAVAS2 (Shi et al. Citation2019) and then edited using Apollo (Misra and Harris Citation2006). The genome sequence and annotations have been deposited in GenBank with accession numbers MT830857.
The cp genome of A. scoparia from China is characterized by a typical circular DNA molecule with a length of 151,008 bp, which is slightly shorter than that of the previous one from Pakistan. This genome consists of a large single copy (LSC) region, a small single copy (SSC) region, and a pair of inverted repeat (IRs) regions, with lengths of 82,778 bp, 18,306 bp, and 24,962 bp, respectively. The overall GC content of the total genome, LSC, SSC, and IR regions are 37.47%, 35.56%, 30.75%, and 43.08%, respectively, which is similar to other taxa in Artemisia (Kim et al. Citation2020). Besides, the genome encodes a total of 132 genes, including 87 protein-coding genes, 37 tRNA genes and 8 rRNA genes, respectively. Overall, the genome is highly similar to those previously published (Iram et al. Citation2019).
The most obvious difference between the two reported genomes is the opposite orientation of the SSC regions. Furthermore, we used DnaSP v6.0 (Rozas et al. Citation2017) software to detect the differences between the two genomes. A total of 79 single nucleotide polymorphisms (SNPs) and 39 insertions and deletions (InDels) were detected. Most SNPs were detected in the LSC (51) and SSC (18) regions, only 10 were found in the IR regions. Among the protein-coding genes, the largest numbers of SNPs were found in gene psaB (9) and ycf1 (7), resulting in the changes of one (452nd, Q-P) and five amino acids (487th, Q-K; 572nd, H-D; 1376th, P-Q; 1475th, M-L; 1552nd, T-N), respectively. For the InDels, nearly half (19 out of 39) involve only single nucleotide, and all InDels were found in the SSC region. A total of four InDels have length over 10 bp, and they were detected in the non-coding regions, including trnR-UCU-trnS-CGA (17 bp), ndhC-trnM-CAU (11 bp), psaJ-rpl33 (15 bp) and petD-intron (20 bp). Our results show that, the four intergenic regions and two protein-coding genes (psaB and ycf1) are hypervariable. The molecular markers based in these regions might be effective in distinguishing A. scoparia below the species level.
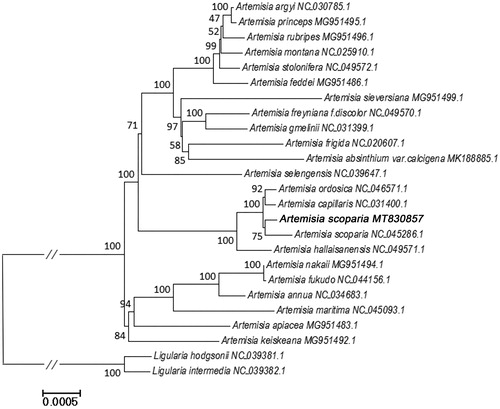
Finally, we inferred phylogenetic relationships among 22 Artemisia species based on the complete cp genome sequences. Two Ligularia species were used as outgroups. The cp genome sequences of 24 species were downloaded from GenBank (the accession numbers are shown in ). All cp genome sequences were aligned using MAFFT (v 7.450) (Rozewicki et al. Citation2019). The phylogenetic tree was constructed using the Maximum Likelihood (ML) method implemented in RaxML (v8.2.4) (Stamatakis Citation2014). The bootstrap analysis was performed with 1,000 replicates. In the phylogenetic tree, most nodes have high bootstrap support values that are > 70. From , the two A. scoparia species are clustered together, as expected. And they are closely related to A. ordosica and A. capillaris. The current study showed that there are significant differences between the cp genomes of the two A. scoparia geotypes. Futhure studies are needed to determine whether there are chemical and pharmacological differences between them.
Figure 1. Phylogenetic relationships of Artemisia species inferred using Maximum likelihood (ML) method. The phylogenetic tree constructed using the complete chloroplast genome sequences among the 25 samples. Numbers near the nodes represent bootstrap values. Relative branch lengths are indicated. Bootstrap values were calculated from 1000 replicates. Two taxa, namely, L. hodgsonii and L. intermedia were used as outgroup taxa.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/nuccore/MT830857. The voucher sample has been deposited in the Herbarium of Institute of Medicinal Plant Development in Beijing, China with the accession number: implad201910001.
Additional information
Funding
References
- Abad MJ, Bedoya LM, Apaza L, Bermejo P. 2012. The artemisia L. Genus: a review of bioactive essential oils. Molecules. 17(3):2542–2566.
- Boudreau A, Richard AJ, Burrell JA, King WT, Dunn R, Schwarz JM, Ribnicky DM, Rood J, Salbaum JM, Stephens JM. 2018. An ethanolic extract of Artemisia scoparia inhibits lipolysis in vivo and has antilipolytic effects on murine adipocytes in vitro. Am J Physiol Endocrinol Metab. 315(5):e1053–e1061.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18–e18.
- Geng CA, Huang XY, Chen XL, Ma YB, Rong GQ, Zhao Y, Zhang XM, Chen JJ. 2015. Three new anti-HBV active constituents from the traditional Chinese herb of Yin-Chen (Artemisia scoparia). J Ethnopharmacol. 176:109–117.
- Iram S, Hayat MQ, Tahir M, Gul A, Ahmed I. 2019. Chloroplast genome sequence of Artemisia scoparia: comparative analyses and screening of mutational hotspots. Plants (Basel, Switzerland). 8(11):476.
- Kim GB, Lim CE, Kim JS, Kim K, Lee JH, Yu HJ, Mun JH. 2020. Comparative chloroplast genome analysis of Artemisia (Asteraceae) in East Asia: insights into evolutionary divergence and phylogenomic implications. BMC Genomics. 21(1):415.
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10 (3):R25.
- Misra S, Harris N. 2006. Using Apollo to browse and edit genome annotations. Curr Protoc Bioinformatics. 12:9.5.1-9.5.28.
- Nam SY, Han NR, Rah SY, Seo Y, Kim HM, Jeong HJ. 2018. Anti-inflammatory effects of Artemisia scoparia and its active constituent, 3,5-dicaffeoyl-epi-quinic acid against activated mast cells. Immunopharmacol Immunotoxicol. 40 (1):52–58.
- Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A. 2017. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 34(12):3299–3302.
- Rozewicki J, Li S, Amada KM, Standley DM, Katoh K. 2019. MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Res. 47 (W1):W5–w10.
- Sajid M, Rashid Khan MR, Shah NA, Waris TS, Younis T, Ullah S, Ahmed N. 2017. Evaluation of Artemisia scoparia for hemostasis promotion activity. Pak J Pharm Sci. 30(5):1709–1713.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47 (W1):W65–w73.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
