Abstract
The black dead leaf butterfly Doleschallia melana (Staudinger, 1886) (Nymphalidae) is a leaf mimic endemic to Indonesia. Genome skimming by Illumina sequencing enabled the assembly of a complete 15,269 bp circular mitogenome from D. melana consisting of 80.0% AT nucleotides, 22 tRNAs, 13 protein-coding genes, 2 rRNAs and a control region in the typical butterfly gene order. Doleschallia melana COX1 features an atypical CGA start codon while ATP6, COX1, COX2, ND1, ND2, ND4, and ND5 exhibit incomplete stop codons. Phylogenetic reconstruction places D. melana as the sister taxon to the clade containing the tribes Kallimini, Nymphalini and Junoniini.
The leaf-mimicking genus Doleschallia (Suzuki et al. Citation2014) was initially classified within the ‘dead leaf butterflies’ in nymphalid tribe Kallimini (Wahlberg et al. Citation2005). However, recent molecular data suggests that Doleschallia is most closely related to tribe Melitaeini and not found within a monophyletic Kallimini (Su et al. Citation2017). Doleschallia melana, endemic to Indonesia, has received little research attention since its original description (Staudinger and Schatz Citation1886). Here, we report the complete mitochondrial genome sequence of D. melana from specimen Dolmel2016.1 that was collected on Seram Island, Indonesia (GPS 3.133S, 129.500E) in June 2016 and has been pinned, spread, and deposited in the Wallis Roughley Museum of Entomology, University of Manitoba (voucher WRME0507729).
DNA was prepared (McCullagh and Marcus Citation2015) and sequenced by Illumina NovaSeq6000 (San Diego, CA) (Marcus Citation2018). The mitogenome of D. melana (Genbank MT704829) was assembled and annotated with Geneious 11.0.4 from 16,411,934 paired 151 bp reads using a Junonia stygia (Lepidoptera: Nymphalidae) reference mitogenome (MN623383, Living Prairie Mitogenomics Consortium Citation2019). Annotation was in reference to Junonia stygia (MG623383, Living Prairie Mitogenomics Consortium Citation2019). The D. melana nuclear rRNA repeat (Genbank MT704832) was assembled and annotated using Junonia stygia (MN623382) (Living Prairie Mitogenomics Consortium Citation2019) and Precis andremiaja (Lepidoptera: Nymphalidae, MH917708) (Lalonde and Marcus Citation2019) reference sequences.
The D. melana circular 15,269 bp mitogenome assembly was composed of 130,494 paired reads with nucleotide composition 40.3% A, 12.4% C, 7.6% G, and 39.8% T. The gene composition and order in D. melana matches other butterfly mitogenomes (McCullagh and Marcus Citation2015; Living Prairie Mitogenomics Consortium Citation2019; Lalonde and Marcus Citation2019). Doleschallia melana COX1 features an atypical CGA start codon like many other insects (Liao et al. Citation2010). The mitogenome contains five protein-coding genes (COX1, COX2, ND2, ND4, ND5) with single-nucleotide (T––) stop codons and two protein coding genes (ATP6, ND1) with two-nucleotide (TA) stop codons completed by post-transcriptional addition of 3′-A residues. The locations and structures of tRNAs were determined using ARWEN v.1.2 (Laslett and Canback Citation2008). tRNAs have typical cloverleaf secondary structures except that the dihydrouridine arm is replaced by a loop in trnS (AGN), while the mitochondrial rRNAs and control region are typical for Lepidoptera (McCullagh and Marcus Citation2015).
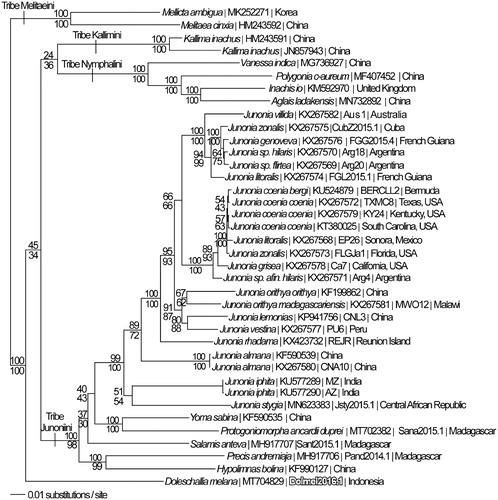
We reconstructed a phylogeny using complete mitogenomes from D. melana and 37 mitogenomes from subfamily Nymphalinae (McCullagh and Marcus Citation2015; Peters and Marcus Citation2016; Peters and Marcus Citation2017; Lalonde and Marcus CitationSubmitted). Mitogenome sequences were aligned in CLUSTAL Omega (Sievers et al. Citation2011), then analyzed in PAUP* 4.0b8/4.0d78 (Swofford Citation2002) by parsimony and maximum likelihood (model selected by jModeltest 2.1.7) (Darriba et al. Citation2012), followed by likelihood ratio test (Huelsenbeck and Rannala Citation1997) (). Doleschallia melana was placed outside tribe Kallimini as the basal lineage among the tribes Nymphalini, Junoniini and Kallimini, contrary to some early phylogenetic analyses (Wahlberg et al. Citation2005; Nylin and Wahlberg Citation2008), but consistent with more recent analyses which used maximum likelihood and Bayesian phylogenetics (Wahlberg et al. Citation2009; Su et al. Citation2017).
Figure 1. Maximum likelihood phylogeny (GTR + G model, G = 0. 2250, likelihood score 101051.99990) of Doleschallia melana and 37 other mitogenomes from subfamily Nymphalinae based on 1 million random addition heuristic search replicates (with tree bisection and reconnection). One million maximum parsimony heuristic search replicates produced 8 trees (parsimony score 16,612 steps) which differed only in the arrangement of three Junonia coenia mitogenome. One parsimony tree had an identical topology to the maximum likelihood tree depicted here. Numbers above each node are maximum likelihood bootstrap values and numbers below each node are maximum parsimony bootstrap values (each from 1 million random fast addition search replicates).
Acknowledgements
We thank Mackenzie Alexiuk and Josephine Payment for their constructive criticism on this manuscript and Genome Quebec for assistance with library preparation and sequencing.
Disclosure statement
The authors report no conflicts of interest and are solely responsible for this paper.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference numbers MT704829 and MT704832.
Additional information
Funding
References
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Huelsenbeck JP, Rannala B. 1997. Phylogenetic methods come of age: testing hypotheses in an evolutionary context. Science. 276(5310):227–232.
- Lalonde MML, Marcus JM. 2019. The complete mitochondrial genome of the Madagascar banded commodore butterfly Precis andremiaja (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA B Resour. 4(1):277–279.
- Lalonde MML, Marcus JM. Submitted. The complete mitochondrial genome of the Malagasy clouded mother-of-pearl butterfly Protogoniamorpha ancardii duprei (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA B Resour. Submitted June 2020. doi:10.1080/23802359.2020.1810156
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Liao F, Wang L, Wu S, Li Y-P, Zhao L, Huang G-M, Niu C-J, Liu Y-Q, Li M-G. 2010. The complete mitochondrial genome of the fall webworm, Hyphantria cunea (Lepidoptera: Arctiidae). Int J Biol Sci. 6(2):172–186.
- Living Prairie Mitogenomics Consortium. 2019. The complete mitochondrial genome of the brown pansy butterfly, Junonia stygia (Aurivillius, 1894), (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA Part B. 5:41–43.
- Marcus JM. 2018. Our love-hate relationship with DNA barcodes, the Y2K problem, and the search for next generation barcodes. AIMS Genet. 5(1):1–23.
- McCullagh BS, Marcus JM. 2015. The complete mitochondrional genome of Lemon Pansy, Junonia lemonias (Lepidoptera: Nymphalidae: Nymphalinae. J Asia-Pacific Ent. 18(4):749–755.
- Nylin S, Wahlberg N. 2008. Does plasticity drive speciation? Host-plant shifts and diversification in Nymphalinae butterflies (Lepidoptera: Nymphalidae) during the tertiary. Biol J Linn Soc. 94(1):115–130.
- Peters MJ, Marcus JM. 2016. The complete mitochondrial genome of the Bermuda buckeye butterfly Junonia coenia bergi (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA B Resour. 1(1):739–741.
- Peters MJ, Marcus JM. 2017. Taxonomy as a hypothesis: testing the status of the Bermuda buckeye butterfly Junonia coenia bergi (Lepidoptera: Nymphalidae). Syst Entomol. 42(1):288–300.
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539.
- Staudinger O, Schatz E. 1886. Doleschallia Feld. Exot Schmett. 1:104.
- Su C, Shi Q, Sun X, Ma J, Li C, Hao J, Yang Q. 2017. Dated phylogeny and dispersal history of the butterfly subfamily Nymphalinae (Lepidoptera: Nymphalidae). Sci Rep. 7(1):8799.
- Suzuki TK, Tomita S, Sezutsu H. 2014. Gradual and contingent evolutionary emergence of leaf mimicry in butterfly wing patterns. BMC Evol Biol. 14:229.
- Swofford DL. 2002. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sunderland, MA: Sinauer Associates.
- Wahlberg N, Brower AVZ, Nylin S. 2005. Phylogenetic relationships and historical biogeography of tribes and genera in the subfamily Nymphalinae (Lepidoptera: Nymphalidae). Biol J Linn Soc. 86(2):227–251.
- Wahlberg N, Leneveu J, Kodandaramaiah U, Peña C, Nylin S, Freitas AVL, Brower AVZ. 2009. Nymphalid butterflies diversify following near demise at the Cretaceous/Tertiary boundary. Proc Biol Sci. 276(1677):4295–4302.
