Abstract
In this study, the complete mitochondrial genomes of Myzornis pyrrhoura and Fulvetta vinipectus were sequenced and described for the first time. The whole mitochondrial genomes of M. pyrrhoura and F. vinipectus are 17,397 bp and 16,961 bp in length, with the G + C percentage 46.34% and 47.36%, respectively. Both genomes contain 13 protein-coding genes, 22 transfer RNA genes, 2 ribosome RNA genes, and 1 non-coding control region. The arrangement of genes is identical to mitochondrial genomes of Sylviidae species reported previously. A phylogenetic reconstruction supported that M. pyrrhoura and F. vinipectus are members of family Sylviidae. The mitochondrial genomes of these two species reported here would be helpful in better understanding the phylogeny and evolution of Sylviidae.
Introduction
The fire-tailed Myzornis (Myzornis pyrrhoura) and white-browed fulvetta (Fulvetta vinipectus) (Aves, Passeriformes, Sylviidae) are two species of small-sized sylviid babblers, which are both rated Least Concern on the IUCN Red List (IUCN Citation2018). Myzornis pyrrhoura has bright green color with black mask and crown scalloping, having red flash in black-and-white wing, red-sided tail, and rather long, thin, slightly decurved black bill (Josep et al. Citation2007). Fulvetta vinipectus is a small fulvetta in soft rich browns and ochrous-buffs with pale eye in dark mask and bold white supercilium (Josep et al. Citation2007). They are both resident birds and sympatric breeders above 2700 m in Gaoligong Mountains (Wang et al. Citation2016; Liang et al. Citation2017). The samples were collected around Pianma Pass (25°58′–26°03′N, 98°41′–98°44′E; 3000–3800 m), which is located in the middle section of Gaoligong Mountains (Liang et al. Citation2017). The two specimens M. pyrrhoura and F. vinipectus are deposited in the Zoological Collection Center of Southwest Forestry University, under the voucher number of SWFU-b0466 and SWFU-b0470.
Methods
The total mitochondrial DNA was extracted from the muscle tissue and was sequenced using next-generation sequencing. The entire mitochondrial genome sequences of M. pyrrhoura and F. vinipectus have been deposited under the GenBank accession numbers MK612108 and MT263986.
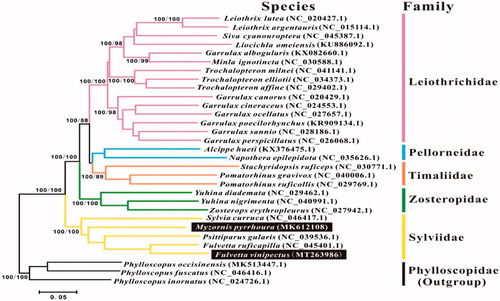
Phylogenetic tree of these two species among babblers were presented by maximum parsimony (MP) analyses using PAUP 4.0b10 software with 1000 bootstrap replication (Swofford Citation2002). Bayesian inference was calculated with MrBayes3.1.2. A general time-reversible (GTR) model of DNA substitution (Rodríguez et al. Citation1990) and a proportion of invariant sites and unequal rates among sites were modeled with a gamma distribution (GTR + I+G) (Yang Citation1996; Ronquist and Huelsenbeck Citation2003). Four Markov chains were run for 2,000,000 generations until the average standard deviation (SD) was less than 0.01. Sequences of other babblers including Pomatorhinus gravivox, Stachyridopsis ruficeps, and Pomatorhinus ruficollis, etc., were obtained from GenBank (). Three Phylloscopidae species were used as the outgroup following Cai et al. (Citation2019) and Gelang et al. (Citation2009).
Figure 1. The phylogenetic tree based on combined protein-coding gene sequences of F. vinipectus and M. pyrrhoura and 26 relative species. The numbers following the species names are GenBank accession numbers. Numbers at a node refer to maximum parsimony (BP, left) and Bayesian posterior probability (BPP, right).
Results
The complete mitochondrial genomes of M. pyrrhoura and F. vinipectus are 17,397 bp and 16,961 bp in length, respectively. The base composition of M. pyrrhoura mitochondrial genome G + C percentage was 46.34% and F. vinipectus 47.36%. Each of them consists of 13 protein coding, two rRNA, 22 tRNA genes, and two non-coding control regions (D-loop). The components and arrangement of genes were the same as those of other babblers species reported previously, such as Garrulax affinis (Huang et al. Citation2016) and Phylloscopus inornatus (Qing et al. Citation2015). For both M. pyrrhoura and F. vinipectus, COI and ND3 are initiated with ‘ATT’ and ‘ATA’, respectively. While the remaining 11 protein-coding genes begin with an ‘ATG’ start codon. Protein-coding genes of M. pyrrhoura were stopped by four types of complete stop codons (TAA for ND1, ND2, ND3, ND4L, COII, ATP6 and ATP8, AGA for ND5, AGG for COI, TAG for Cytb and ND6) and one incomplete stop codon (T for COIII and ND4), while protein-coding genes of F. vinipectus were stopped by the same five types of complete stop codons but with different arrangement (TAA for ND2, ND3, ND4L, ND5, Cytb, COII, ATP6 and ATP8, AGA for ND1, AGG for COI, TAG for ND6) and one incomplete stop codon (T for COIII and ND4). ND6 and eight tRNA genes, i.e. tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAPro, and tRNAGlu, were encoded on a light strand (L-strand), while all of the remaining genes were located on the heavy strand (H-strand). The two control regions of mitochondrial genome were D-loop1 (1154 bp in M. pyrrhoura and 1144 bp in F. vinipectus) and D-loop2 (657 bp in M. pyrrhoura and 245 bp in F. vinipectus).
The reconstructed phylogenetic tree supported the placement of M. pyrrhoura and F. vinipectus in the Sylviidae family (). All of the nodes were strongly supported by the MP and Bayesian analysis. This study identified F. vinipectus as a close relative of F. ruficapilla, which was also supported by BI analysis results of Cai et al. (Citation2019). Nevertheless, M. pyrrhoura is an unpaired branch separating from the other remaining species in the Sylviidae family, that was also consistent with the result of Cai et al. (Citation2019) and Gelang et al. (Citation2009). The mitochondrial genomes reported here would be useful in the current understanding of the phylogeny and evolution of Sylviidae.
Acknowledgements
Thanks are given to Yuan Li and Junzhu Chen for offering assistance on submitting mitochondrial genome sequences to GenBank. We are also grateful to Xuemei Ding who helped with Bayesian inference.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The entire mitochondrial genome sequences of M. pyrrhoura and F. vinipectus have been deposited under the GenBank accession numbers MK612108 and MT263986, which are available at https://www.ncbi.nlm.nih.gov.
Additional information
Funding
References
- Cai TL, Cibois A, Alström P, Moyle RG, Kennedy JD, Shao S, Zhang R, Irestedt M, Ericson P, Gelang M, et al. 2019. Near-complete phylogeny and taxonomic revision of the world's babblers (Aves: Passeriformes). Mol Phylogenet Evol. 130:346–356.
- Gelang M, Cibois A, Pasquet E, Olsson U, Alström P, Ericson PGP. 2009. Phylogeny of babblers (Aves, Passeriformes): major lineages, family limits and classification. Zool Scripta. 38(3):225–236.
- Huang R, Zhou YY, Yao YF, Zhao B, Zhang YL, Xu HL. 2016. Complete mitochondrial genome and phylogenetic relationship analysis of Garrulax affinis (Passeriformes, Timaliidae). Mitochondrial DNA Part A. 27(5):3502–3503.
- IUCN. 2018. The IUCN red list of threatened species. Version 2018-1; [accessed 2018 Nov 20]. www.incnredlist.org.
- Josep DH, Nigel JC, Norman A, Walter JB, Andrew E, David C. 2007. Handbook of the birds of the world. Picathartes to tits and chickadees. Vol. 12. Barcelona, Spain: Lynx Edicions.
- Liang D, Gao G, Han LX, Luo X. 2017. Breeding biology of fire-tailed Myzornis (Myzornis pyrrhoura) in an alpine environment in Southwestern China. Wilson J Ornith. 129(3):568–575.
- Liang D, Gao G, Wang B, Wang XY, Chen YX, Wu XR, Zhao C, Jiang DM, Han LX, Luo X. 2015. The diversity and vertical distribution of the birds in Middle Gaoligong Mountain, Yunnan. Sichuan J Zool. 34:930–940.
- Qing H, Liu G, Zhou L, Wang J, Li L, Li B, Olsson U. 2015. Complete mitochondrial genome of yellow-browed warbler Phylloscopus inornatus inornatus (Passeriformes: Sylviidae). Mitochondrial DNA. 26(6):939–932.
- Rodríguez F, Oliver JF, Marín A, Medina JR. 1990. The general stochastic model of nucleotide substitutions. J Theor Biol. 142(4):485–501.
- Ronquist F, Huelsenbeck JP. 2003. Mrbayes3: Bayesian Phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Swofford DL. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4.0b10. Massachusetts: Sinauer Associates.
- Wang XY, Liang D, He YX, Luo X. 2016. Breeding ecology of Alcippe vinipectus at Pianma Pass on Gaoligong Mountain, Yunnan. J Southwest Forest Univ. 36:181–186.
- Yang Z. 1996. Among-site rate variation and its impact on phylogenetic analyses. Trends Ecol Evol. 11(9):367–372.
