Abstract
In this study, the complete mitochondrial genome of Sterigmatomyces hyphaenes was sequenced by the next-generation sequencing. The complete mitochondrial genome of S. hyphaenes contained 17 protein-coding genes (PCG), 2 ribosomal RNA (rRNA) genes, and 23 transfer RNA (tRNA) genes. The total size of the S. hyphaenes mitochondrial genome is 26,198 bp, and the GC content of the mitochondrial genome is 42.08%. Phylogenetic analysis based on the combined mitochondrial gene dataset indicated that the mitochondrial genome of S. hyphaenes exhibited a close relationship with that of Rhodotorula mucilaginosa.
The genus Sterigmatomyces was established by Fell for nonfilamentous, yeast-like fungi, which are characterized by a unique method of cell division (Fell Citation1966). The yeast cells produce one or more sterigmata, each of which gives rise to a single conidium (Gueho et al. Citation1990; Messner et al. Citation1994). Several species have been described in this genus (Sonck Citation1969; Rodrigues de Miranda Citation1975). Some species of this genus have excellent salt tolerance, and some species can produce lactosucrose (Lee et al. Citation2007; Al-Tohamy et al. Citation2020). Limited morphological characteristics make it difficult to identify or classify Sterigmatomyces species accurately only according to morphology (Gueho et al. Citation1990; Messner et al. Citation1994). Mitochondrial genome has been widely used in the phylogeny of basidiomycete species (Wang et al. Citation2020; Li, He et al. Citation2020). However, up to now, no mitochondrial genome from the genus Sterigmatomyces has been published, and the complete mitochondrial genome of Sterigmatomyces hyphaenes reported here will promote the understanding of the phylogeny and taxonomy of this fungal group.
The specimen (S. hyphaenes) was collected from Sichuan, China (103.26 E; 30.55 N), and was stored in the Culture Collection Center of Chengdu University (No. Asas_ca01). The complete mitochondrial genome of S. hyphaenes was sequenced and de novo assembled according to previously described methods (Li, Liao et al. Citation2018; Li, Xiang et al. Citation2019; Wang et al. Citation2020). Briefly, we extracted the total genomic DNA of S. hyphaenes using a Fungal DNA Kit D3390-00 (Omega Bio-Tek, Norcross, GA). And then we purified the extracted genomic DNA using a Gel Extraction Kit (Omega Bio-Tek, Norcross, GA). The purified DNA was stored in Chengdu University (No. DNA_Asas_ca01). Sequencing libraries were constructed using a NEBNext® Ultra™ II DNA Library Prep Kit (NEB, Beijing, China). Whole genomic sequencing (WGS) of S. hyphaenes was conducted using the Illumina HiSeq 2500 Platform (Illumina, SanDiego, CA). We de novo assembled the mitochondrial genome of S. hyphaenes using SPAdes 3.9.0 (Bankevich et al. Citation2012; Li, Ren et al. Citation2020). The complete mitochondrial genome of S. hyphaenes was annotated according to the previous described methods (Li, Chen et al. Citation2018; Li, Wang et al. Citation2018).
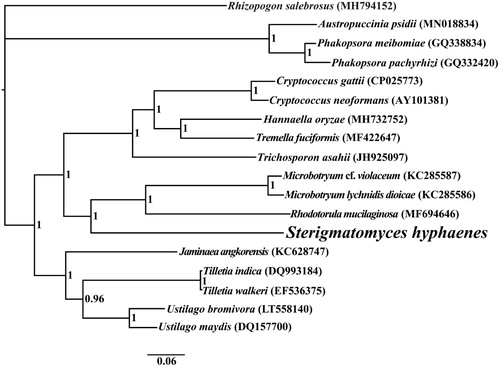
The complete mitochondrial genome of S. hyphaenes is 26,198 bp in length, with the base composition as follows: A (28.45%), T (29.48%), G (19.73%), and C (22.35%). The complete mitochondrial genome of S. hyphaenes contains 17 protein-coding genes, 2 ribosomal RNA genes (rns and rnl), and 23 transfer RNA (tRNA) genes. To investigate the phylogenetic status of the mitogenome of S. hyphaenes, we constructed a phylogenetic tree for 18 basidiomycete species. Rhizopogon salebrosus from the Boletales order was set as the outgroup (Li, Ren et al. Citation2019a). The phylogenetic tree was constructed using the Bayesian analysis (BI) method based on the combined 14 core protein-coding genes according to previously described methods (Li, Wang, Jin, Chen, Xiong, Li, Liu et al. Citation2019; Li, Wang, Jin, Chen, Xiong, Li, Zhao et al. Citation2019; Li, Yang et al. Citation2020). As shown in the phylogenetic tree (), the mitochondrial genome of S. hyphaenes exhibited a close relationship with that of Rhodotorula mucilaginosa (Gan et al. Citation2017).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
This mitogenome of S. hyphaenes was submitted to GenBank under the accession number of MT755636. (https://www.ncbi.nlm.nih.gov/nuccore/ MT755636).
Additional information
Funding
References
- Al-Tohamy R, Kenawy ER, Sun J, Ali SS. 2020. Performance of a newly isolated salt-tolerant yeast strain Sterigmatomyces halophilus SSA-1575 for azo dye decolorization and detoxification. Front Microbiol. 11:1163.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Fell JW. 1966. Sterigmatomyces, a new fungal genus from marine areas. Antonie Van Leeuwenhoek. 32(1):99–104.
- Gan HM, Thomas BN, Cavanaugh NT, Morales GH, Mayers AN, Savka MA, Hudson AO. 2017. Whole genome sequencing of Rhodotorula mucilaginosa isolated from the chewing stick (Distemonanthus benthamianus): insights into Rhodotorula phylogeny, mitogenome dynamics and carotenoid biosynthesis. PeerJ. 5:e4030.
- Gueho E, Kurtzman CP, Peterson SW. 1990. Phylogenetic relationships among species of Sterigmatomyces and Fellomyces as determined from partial rRNA sequences. Int J Syst Bacteriol. 40(1):60–65.
- Lee JH, Lim JS, Park C, Kang SW, Shin HY, Park SW, Kim SW. 2007. Continuous production of lactosucrose by immobilized Sterigmatomyces elviae mutant. J Microbiol Biotechnol. 17(9):1533–1537.
- Li Q, Chen C, Xiong C, Jin X, Chen Z, Huang W. 2018. Comparative mitogenomics reveals large-scale gene rearrangements in the mitochondrial genome of two Pleurotus species. Appl Microbiol Biotechnol. 102(14):6143–6153.
- Li Q, He X, Ren Y, Xiong C, Jin X, Peng L, Huang W. 2020. Comparative mitogenome analysis reveals mitochondrial genome differentiation in ectomycorrhizal and asymbiotic Amanita species. Front Microbiol. 11:1382.
- Li Q, Liao M, Yang M, Xiong C, Jin X, Chen Z, Huang W. 2018. Characterization of the mitochondrial genomes of three species in the ectomycorrhizal genus Cantharellus and phylogeny of Agaricomycetes. Int J Biol Macromol. 118:756–769.
- Li Q, Ren Y, Shi X, Peng L, Zhao J, Song Y, Zhao G. 2019. Comparative mitochondrial genome analysis of two ectomycorrhizal fungi (Rhizopogon) reveals dynamic changes of intron and phylogenetic relationships of the subphylum agaricomycotina. IJMS. 20(20):5167.
- Li Q, Ren Y, Xiang D, Shi X, Zhao J, Peng L, Zhao G. 2020. Comparative mitogenome analysis of two ectomycorrhizal fungi (Paxillus) reveals gene rearrangement, intron dynamics, and phylogeny of basidiomycetes. IMA Fungus. 11:12.
- Li Q, Wang Q, Chen C, Jin X, Chen Z, Xiong C, Li P, Zhao J, Huang W. 2018. Characterization and comparative mitogenomic analysis of six newly sequenced mitochondrial genomes from ectomycorrhizal fungi (Russula) and phylogenetic analysis of the Agaricomycetes. Int J Biol Macromol. 119:792–802.
- Li Q, Wang Q, Jin X, Chen Z, Xiong C, Li P, Liu Q, Huang W. 2019. Characterization and comparative analysis of six complete mitochondrial genomes from ectomycorrhizal fungi of the Lactarius genus and phylogenetic analysis of the Agaricomycetes. Int J Biol Macromol. 121:249–260.
- Li Q, Wang Q, Jin X, Chen Z, Xiong C, Li P, Zhao J, Huang W. 2019. Characterization and comparison of the mitochondrial genomes from two Lyophyllum fungal species and insights into phylogeny of Agaricomycetes. Int J Biol Macromol. 121:364–372.
- Li Q, Xiang D, Wan Y, Wu Q, Wu X, Ma C, Song Y, Zhao G, Huang W. 2019. The complete mitochondrial genomes of five important medicinal Ganoderma species: features, evolution, and phylogeny. Int J Biol Macromol. 139:397–408.
- Li Q, Yang L, Xiang D, Wan Y, Wu Q, Huang W, Zhao G. 2020. The complete mitochondrial genomes of two model ectomycorrhizal fungi (Laccaria): features, intron dynamics and phylogenetic implications. Int J Biol Macromol. 145:974–984.
- Messner R, Prillinger H, Altmann F, Lopandic K, Wimmer K, Molnar O, Weigang F. 1994. Molecular characterization and application of random amplified polymorphic DNA analysis of Mrakia and Sterigmatomyces species. Int J Syst Bacteriol. 44(4):694–703.
- Rodrigues de Miranda L. 1975. Two new species of the genus Sterigmatomyces. Antonie Van Leeuwenhoek. 41(2):193–199.
- Sonck CE. 1969. A new yeast species, Sterigmatomyces elviae, isolated from eczematous skin lesions in two patients. Antonie Van Leeuwenhoek. 35(Suppl):E25–6.
- Wang X, Song A, Wang F, Chen M, Li X, Li Q, Liu N. 2020. The 206 kbp mitochondrial genome of Phanerochaete carnosa reveals dynamics of introns, accumulation of repeat sequences and plasmid-derived genes. Int J Biol Macromol. 162:209–219.