Abstract
Dendrobium is one of the largest genera in Orchidaceae with approximately 1500 species. Many of them have been used as Traditional Chinese Medicine (TCM) for hundreds of years. Here, the first complete chloroplast genome sequence of D. densiflorum was reported and characterized. The complete cpDNA of D. densiflorum is a circular molecule of 153,122 bp, which contains 76 protein-coding genes, 30 tRNA genes, and four rRNA genes. Our phylogenetic analysis indicated that the newly sequenced cpDNA of D. densiflorum could be used for the phylogenetic study of Dendrobium species.
Chloroplast genomes (cpDNAs) of seed plants, in general, have relatively small sizes, conserved gene contents, dense coding regions and slower evolutionary rates as compared to nuclear and mitochondrial genomes (Wolfe et al. Citation1987). These unique features mentioned above have led the cpDNA be a focus of research in plant molecular evolution and systematics (Jansen et al. Citation2007). Dendrobium, a genus belongs to the tribe Dendrobieae (Orchidaceae: Epidendroideae), is one of the largest genera in Orchidaceae with approximately 1500 species mainly distributed in tropical Asia, Australasia, and Australia. Because of their beautiful and long-lasting flowers, Dendrobium species are valuable commercially. Moreover, many species in this genus have been used as Traditional Chinese Medicine (TCM) for hundreds of years due to their excellent medicinal merits, i.e., D. densiflorum. Recently, the cpDNA sequences are available for more than 30 Dendrobium species; however, there still none information about the chloroplast genome sequence of D. densiflorum. (Luo et al. Citation2014). Therefore, to provide more insight into the cpDNA structure and evolution study of D. densiflorum, we sequenced the complete cpDNA sequence and reported the cpDNA features of D. densiflorum.
In this study, the total genomic DNA was extracted from 2 g fresh leaves of D. densiflorum (voucher number: LW20190816), and sequenced on Illumina Hiseq 4000 sequencer. The raw reads were trimmed and mapped to the reference cpDNA sequence of D. officinale (NC_024019) according to the method in Niu et al. (Citation2017). The gaps and boundaries of IRs were confirmed by PCR assays. The complete cpDNA sequence has been submitted to DDBJ under the accession number of LC528135.
The complete cpDNA of D. densiflorum is a circular molecule of 152,110 bp. The GC content is 37.13%. It has a typical quartered structure with a pair of inverted repeats (IRs) (26,124 bp each) separated by a large single copy (LSC) (85,078 bp) and a small single copy (SSC) (14,785 bp) regions. The GC content of LSC, SSC and IR were 34.95%, 30.27% and 43.30%, respectively. A total of 111 unique genes were successfully annotated, including 76 protein-coding genes, 30 tRNA genes, and four rRNA genes. Among the eleven ndh genes, only ndhB genes in IR regions were functional with full reading frames, whereas others were truncated or completely lost.
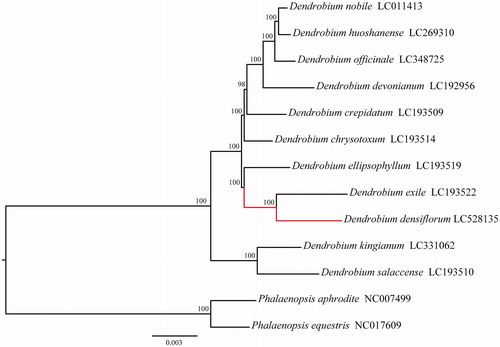
We constructed the maximum-likelihood (ML) tree of the whole cpDNA sequences of 11 Dendrobium species using RAxML 8.0.2 (Stamatakis Citation2014) with Phalaenopsis aphrodite (NC_007499) and Phalaenopsis equestris (NC_017609) as outgroups. As shown in , the ML tree has revealed a sister relationship between D. densiflorum and D. exile, which was consisting with previously reported results (Xiang et al. Citation2013), which indicated that the newly sequenced cpDNA of D. densiflorum could be used for the phylogenetic study of Dendrobium species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 104(49):19369–19374.
- Luo J, Hou BW, Niu ZT, Liu W, Xue QY, Ding XY. 2014. Comparative chloroplast genomes of photosynthetic orchids: insights into evolution of the Orchidaceae and development of molecular markers for phylogenetic applications. PLoS One. 9(6):e99016
- Niu Z, Zhu S, Pan J, Li L, Sun J, Ding X. 2017. Comparative analysis of Dendrobium plastomes and utility of plastomic mutational hotspots. Sci Rep. 7:2073.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wolfe KH, Li WH, Sharp PM. 1987. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA. 84(24):9054–9058.
- Xiang X-G, Schuiteman A, Li D-Z, Huang W-C, Chung S-W, Li J-W, Zhou H-L, Jin W-T, Lai Y-J, Li Z-Y, et al. 2013. Molecular systematics of Dendrobium (Orchidaceae, Dendrobieae) from mainland Asia based on plastid and nuclear sequences. Mol Phylogenet Evol. 69(3):950–960.