Abstract
Alpinia chinensis (Retz.) Rosc is one of Chinese tradition herbal medicine and edible plant in China. In this report, we sequenced the complete chloroplast genome of A. chinensis. Through the assembly annotation of genome with high-throughput sequencing data, which help us to research the evolution. The length of chloroplast sequences was 163,590 bp with a large single-copy region (LSC) and a small single-copy region (SSC), also, two inverted repeat region A (IR), whose length was 88,951, 15,299, and 29,670 bp, respectively. A total of 138 genes were predicted in the complete chloroplast genome, with 36.4% GC content, including 93 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. From the phylogenetic analysis, we could conclude that A. chinensis (Retz.) Rosc. was close to Alpinia oxyphylla in Zingiberaceae.
Alpinia chinensis belongs to Zingiberaceae family, widely distributed in southeast to southwest provinces of China, which is a famous national medicinal with multiple medicinal properties (Qi et al. Citation2020). Alpinia chinensis (Retz.) contain abundant chemical composition, of which most are medical resource such as volatile oils, in the roots, leaves, and seeds. The chloroplast (cp) is an important organelle in plant cells, which can produce essential energy through photosynthesis and has its own independent genetic material (Daniell et al. Citation2016). The cp genome provides valuable information for species identification and phylogenetic analysis (Shen et al. Citation2017). In recent years, researchers have reported the cp genome of medicinal plants. Whereas, the cp genome of A. chinensis has not been reported.
In this study, the complete cp genome of A. chinensis was reported. The leaves of A. chinensis were collected at the Fengwan town, Shaoguan city, Guangdong province, China (E113°51′ 26.9496″, N24°48′42.2712″). The samples were frozen by liquid nitrogen and stored at −80 °C refrigerator before extracting genomic DNA using plant genomic DNA kit (Omega, USA), then constructed DNA library with an average insert size 350-bp using VAHTS Universal DNA Library Prep Kit (Vazyme ND606-01, Nanjing, China), which were kept at Key Laboratory for Crops Genetic Improvement of Guangdong in Guangdong Academy of Agricultural Sciences (specimen code Hsj2020). Finally, the library was sequenced on the Novaseq platform (Illumina, San Diego, CA, USA). The sequence was assembled and annotated by GetOrganelle (Jin et al. Citation2019) and Geseq (Tillich et al. Citation2017), using the cp annotation of Alpinia oxyphylla (KY985237) as a reference. Then, submitted to GenBank with the accession number MT239402.
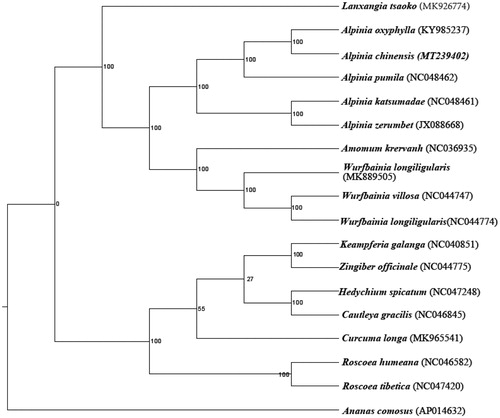
The length of the cp genome sequence was 163,590 bp, with 36.4% GC content, consisted of a large single-copy region (LSC) region (88,951 bp) and small single-copy region (SSC) region (15,299 bp), separated by a pair of inverted repeat region (IR) region (29,670 bp). A total of 138 genes were predicted in the cp genome of A. chinensis, including 93 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Through RaxML software, the complete chloroplast genomes of A. chinensis and other 18 species were constructed by the maximum likelihood method using the GTRGAMMA nucleotide substitution model (Stamatakis Citation2014). So in , We could conclude that A. chinensis was close to A. oxyphylla in Zingiberaceae. These researches will provide valuable genomic information for the barcode and make an essential foundation for molecular breeding and genetic engineering of A. chinensis in the future.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are openly available in [GenBank] at [https://www.ncbi.nlm.nih.gov/nuccore/MT239402.1], reference number [MT239402.1].
Additional information
Funding
References
- Daniell H, Lin Choun S, Yu M, Chang Wan J. 2016. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17(1):1–29.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2019. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. BioRxiv. 256479.
- Qi Y, Cheng X, Jing H, Yan T, Xiao F, Wu B, Bi K, Jia Y. 2020. Comparative pharmacokinetic study of the components in Alpinia oxyphylla Miq.– Schisandra chinensis (Turcz.) Baill. herb pair and its single herb between normal and Alzheimer's disease rats by UPLC-MS/MS. J Pharm Biomed Anal. 177:112874.
- Shen X, Wu M, Liao B, Liu Z, Rui B, Xiao S, Li X, Zhang B, Xu J, Chen S. 2017. Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules. 22(8):1330.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.