Abstract
Heteromorpha arborescens has long been recognized and cultivated as an important medicinal plant. We reported its complete plastid genome for the first time and reconstructed its phylogenetic position. The complete plastid genome was 157,172 bp in length with a typical quadripartite organization: a large single-copy (LSC) region of 86,436 bp, a small single-copy (SSC) region of 18,222 bp, and two inverted repeat regions (IRa and IRb), each of 26,257 bp. A total of 130 functional genes were recovered, consisting of 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The phylogenetic analysis suggested that H. arborescens is sister to other species except Cetella arborescens in Apiaceae with strong ultrafast support.
Keywords:
Heteromorpha arborescens (Spreng.) Cham. & Schltdl., a large shrub, small or medium tree species of the Apiaceae family, is an important medicinal plant species distributed in tropical Africa (Maroyi Citation2018). Furthermore, Heteromorpha is a monophyletic taxon based on previous phylogenetic analyses of molecular data (Calviño et al. Citation2006). But, the relationship between taxa in this genus is confusing. Here, we firstly reported the complete plastid genome of H. arborescens which may help in molecular and phylogenetic studies of this plant.
The total genomic DNA was isolated using fresh and healthy leaf tissues with a modified CTAB method (Doyle and Doyle Citation1987). The voucher specimen (S2007) was collected from Nanjing Botanical Garden Mem. Sun Yat-Sen (32°3′31.33″N, 118°49′48.55″E) and deposited in the herbarium of Institute of Botany, Jiangsu Province and Chinese Academy of Sciences (NAS). Paired-end (PE) sequencing was conducted on the Illumina Novaseq platform and sequenced using Illumina genome analyzer (Illumina novaseq6000). We employed GetOrganelle pipeline (Jin et al. Citation2018) and PGA program (Qu et al. Citation2019) to assemble and annotate the plastome, respectively. Geneious Prime 2020.0.5 (Kearse et al. Citation2012) was used to verify the accuracy of the assembly, and to correct the start/stop codons and intron/exon boundaries of the annotation. The annotated plastome was deposited in GenBank (accession number: MT786054).
The complete chloroplast genome of Heteromorpha arborescens was 1,57,172 bp in length with a typical quadripartite organization: a large single-copy (LSC) region of 86,436 bp, a small single-copy (SSC) region of 18,222 bp, and two inverted repeat regions (IRa and IRb), each of 26,257 bp. A total of 130 functional genes were recovered, consisting of 85 protein-coding genes, 37 tRNA genes and 8 rRNA genes. The overall GC content of the whole plastome is 37.9%.
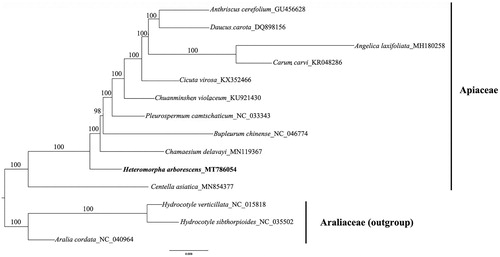
To reconstruct the phylogenetic tree of Heteromorpha arborescens, we included 13 plastid genomes in previous publications in GenBank (). We aligned 78 protein-coding genes using MAFFT v.7.388 (Katoh and Standley Citation2013) with default parameters and manually adjusted misaligned regions using Geneious. We reconstructed the maximum likelihood phylogeny based on a concatenation of the 78 genes building on IQ-tree (Trifinopoulos et al. Citation2016) with the models recommended by ModelFinder (Kalyaanamoorthy et al. Citation2017). Our phylogenetic analysis suggested that H. arborescens is sister to other species except Cetella arborescens in Apiaceae with strong ultrafast support. Additionally, Hydrocotyle species is confirmed in Aralicaceae based on plastome phylogeny, which is consistent with the opinion in the APG IV (Angiosperm Phylogeny Group Citation2016). This study also demonstrated the potential power of genomic data in resolving the phylogenetic relationships among Heteromorpha and other species in Apiaceae.
Data avaliability statement
The data that support the findings of this study are openly avaliable in National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov/, reference number [MT786054].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Angiosperm Phylogeny Group. 2016. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181:1–20.
- Calviño CI, Tilney PM, Wyk BE, Downie SR. 2006. A molecular phylogenetic study of Southern African Apiaceae. Am J Bot. 93(12):1828–1847.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi S, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. DOI:10.1101/256479.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Haeseler AV, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30 (4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Maroyi A. 2018. Heteromorpha arborescens: a review of its botany, medicinal uses, and pharmacological properties. Asian J Pharm Clin Res. 11 (11):75–82.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15 (1):50.
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235.