Abstract
The Weddell seal, Leptonychotes weddellii, which belongs to the family Phocidae, is an abundant pinniped that inhabits the Antarctica. Here, we present the complete mitochondrial genome and phylogeny of L. weddellii. The total length of the mitogenome is 16,762 bp, consisting of 13 protein-coding genes (PCGs), 22 tRNA genes, and 2 rRNA genes. The base composition of the mitogenome is 34.26% (A), 25.51% (T), 27.09% (C), and 13.11% (G), and 40.22% for overall GC contents. The description of this mitogenome can provide information about variations at the intra-species level and aid phylogenetic studies in family Phocidae.
The Weddell seal (Leptonychotes weddellii) is one of the largest phocid seals inhabiting the coastal areas of Antarctica (Wheatley et al. Citation2006). This species shows the southernmost distribution and breeds on ice (Kooyman Citation1981). The breeding population is estimated as stable and is considered as Least Concern by the IUCN (Hückstädt Citation2015). The complete mitochondrial genome information of a species is important to understanding its mitogenomic evolution (Douglas and Gower Citation2010). Although the mitogenomic data of L. weddellii (AM181025) have been reported (Arnason et al. Citation2006), there is no specific information on the sampling location (described as only Antarctica). We tried to identify the intra-species mutation of mitogenome with more accurate geographical information. In this study, we sequenced and analyzed the mitogenome of L. weddellii and compared it with the results of the previous study and those of other species in the family Phocidae.
Here, we assembled the complete mitogenome using Illumina data and revealed the phylogeny of L. weddellii and its relationship with relative species. The mitogenome sequence was deposited in GenBank as MT755639. The carcass of L. weddellii was collected from Terra Nova Bay (74°38′15.69″S, 164°13′43.46″E), Ross Sea, Antarctica, on November 18, 2017. The specimen (proof number: WS01) was stored at the Korea Polar Research Institute (KOPRI), Incheon, South Korea. Total genomic DNA was isolated from muscle tissue using the DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany), and then next-generation sequencing (NGS) was performed. High quality reads (>Q20) of 0.89 Gb were obtained from paired-end (PE) raw reads and were de novo assembled using the CLC genome assembler (v. 4.21, CLC Inc., Aarhus, Denmark) with a reads distance 400–600 bp, minimum length fraction for 0.5, and similarity for 0.8. The mitogenomes from the initially assembled contigs were processed to generate a single draft sequence (Lee et al. Citation2018). Overall sequences were annotated using GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq-app.html) and Artemis annotation tool (Rutherford et al. Citation2000) with NCBI BLASTN search (https://blast.ncbi.nlm.nih.gov). In addition, the mitogenome sequence variation was compared at the intra-species level between a reference mitogenome (AM181025) and a newly assembled mitogenome (MT755639). Two mitogenome sequences were aligned by MAFFT (Katoh and Standley Citation2013) and analyzed using single nucleotide polymorphism (SNP), and insertions and deletions (InDel) were detected. The results revealed 12 SNPs and 9 InDels between the two L. weddellii mitogenomes. Phylogenetic analysis was performed using the concatenated 13 PCGs set of the L. weddellii mitogenome with the inclusion of 16 published mitogenomes from the family Phocidae. A phylogenetic tree was constructed by the maximum likelihood (ML) method with 1000 bootstrap replicates via MEGA 7.0 program (Kumar et al. Citation2016) using the Tamura-Nei model ().
Figure 1. A phylogenetic tree of 14 species of the family Phocidae. Thirteen PCGs in the mitochondrial genome were aligned and used to generate the maximum likelihood phylogenetic tree. The numbers in the nodes indicate bootstrap support values (>50%) from 1,000 replicates.
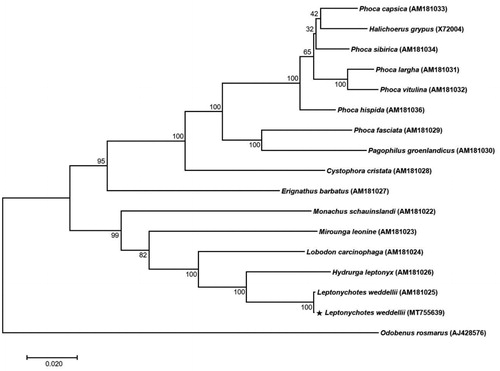
The complete mitochondrial genome sequence of L. weddellii was 16,762 bp in length. A total of 37 genes were predicted in the genome, consisting of 13 protein-coding genes (PCGs), 22 tRNA genes, and 2 rRNA genes. The nucleotide composition of L. weddellii is as follows: 34.26% (A), 25.51% (T), 27.09% (C), and 13.11% (G), and 40.22% for overall GC contents. As expected, both L.weddellii genomes group together with 100% bootstrap in the ML tree.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the finding of this study are openly available in NCBI at (https://www.ncbi.nlm.nih.gov), reference number [MT755639].
Additional information
Funding
References
- Arnason U, Gullberg A, Janke A, Kullberg M, Lehman N, Petrov EA, Väinölä R. 2006. Pinniped phylogeny and a new hypothesis for their origin and dispersal. Mol Phylogenet Evol. 41(2):345–354.
- Douglas DA, Gower DJ. 2010. Snake mitochondrial genomes: phylogenetic relationships and implications of extended taxon sampling for interpretations of mitogenomic evolution. BMC Genomics. 11:14.
- Hückstädt L. 2015. Leptonychotes weddellii. The IUCN Red List of Threatened Species 2015: e.T11696A45226713.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kooyman GL. 1981. Weddell Seal: consummate diver. Cambridge: Cambridge University Press.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lee HO, Choi JW, Baek JH, Oh JH, Lee SC, Kim CK. 2018. Assembly of the mitochondrial genome in the campanulaceae family using Illumina low-coverage sequencing. Genes. 9(8):383.
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics. 16(10):944–945.
- Wheatley K, Bradshaw C, Davis L, Harcourt R, Hindell M. 2006. Influence of maternal mass and condition on energy transfer in Weddell seals. J Anim Ecol. 75(3):724–733.
