Abstract
Phlegmariurus (Herter) Holub is the largest genus of Lycopodiaceae, with about 250 species distributed in the tropics and subtropics of the world. Phlegmariurus phlegmaria is the representative species of Phlegmariurus. In this study, we reported the complete chloroplast genome of P. phlegmaria. This complete chloroplast genome is 1,49,711 bp in size. In total, 134 genes were identified, including 84 protein-coding genes, 42 tRNA genes, and eight rRNA genes. In phylogenetic analysis, a close relationship with genus Huperzia was supported by maximum-likelihood (ML) tree. The complete plastome of P. phlegmaria will provide potential genetic resources to understand the evolution of lycophytes.
Phlegmariurus (Herter) Holub is the largest genus of Lycopodiaceae, with about 250 species distributed in the tropics and subtropics of the world (PPGI Citation2016). Phlegmariurus is mostly closely related to the temperate genus Huperzia (Wikström and Kenrick Citation2000; Field et al. Citation2016), and these two genera are belonging to Lycopodiaceae subfamily Huperzioideae (Wagner and Beitel Citation1992; Øllgaard Citation2015; PPGI Citation2016). Phlegmariurus phlegmaria is the representative species of genus Phlegmariurus. Here, we assembled and characterized the complete plastome of P. phlegmaria. It will provide great genetic resources to amplify the relationship between Phlegmariurus and Huperzia, and understand the phylogeny relationship of lycophytes.
The plant material of P. phlegmaria was collected from city Pingxiang (Guangxi, China; 22.09°N, 106.76°E). Voucher specimen (9461) and DNA sample were deposited in the herbarium of Institute of Botany, CAS (PE). In total, 6G high-quality clean reads (150 bp PE read length) were generated with adaptors trimmed. The Get Organelle (Jin et al. Citation2018), Bandage (Wick et al. Citation2015), GeSeq (Tillich et al. Citation2017), were used to align, assemble, and annotate the chloroplast genome.
The full length of P. phlegmaria chloroplast genome (GenBank Accession No. MT786212) was 1,49,711 bp and comprised a large single-copy region (LSC with 99,862 bp), a small single-copy region (SSC with 19,465 bp), and two inverted repeat regions (IR with 15,192 bp). The plastid genome length of P. phlegmariais similar to P. carinatus and Huperzia javanica (Zhang et al. Citation2017; Luo et al. Citation2019). The overall GC content of P. phlegmaria cp genome was 33.8%, in particular 31.4% in the LSC, 44.0% in the IR, 30.1% in the SSC region. A total of 134 genes were contained in the chloroplast genome, including 84 protein-coding genes (nad5 and ndhF genes were duplicated in the repeat region), 42 tRNA genes (trnA-UGC, trnE-UUC, trnI-GAU, trnN-GUU, trnR-ACG, and trnV-GAC tRNA genes were duplicated in the repeat region), and eight rRNA genes (rrn16, rrn23, rrn4.5, and rrn5 genes were duplicated in the repeat region).
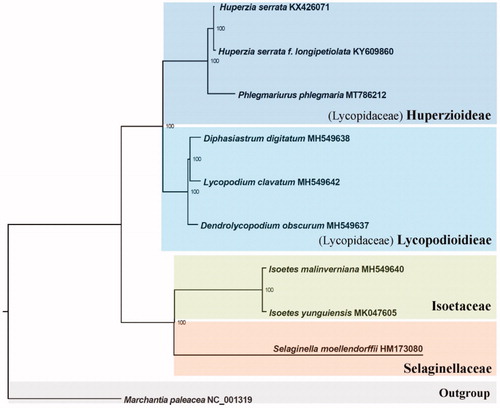
Ten complete chloroplast genomes were selected to infer the phylogenetic relationships among the main representative species of lycophytes with Marchantia paleacea (liverwort) as the out group. All of these ten complete chloroplast sequences were aligned by the MAFFT version 7 software (Katoh and Standley Citation2013) and trimmed by TrimAl (Capella-Gutierrez et al. Citation2009). A maximum-likelihood (ML) tree was inferred by Ultrafast bootstrapping with 1000 replicates through IQ-TREE version 1.5.5 (Nguyen et al. Citation2015) based on the TVM + F + R3 nucleotide substitution model, which was selected by ModelFinder (Kalyaanamoorthy et al. Citation2017). The result revealed that the genus Phlegmariurus is strongly supported as monophyletic and sister to the genus Huperzia. ().
Figure 1. The ML phylogeny results from ten complete plastome sequences by IQ-TREE. Accession numbers: Huperzia serrata KX426071, H. serrata f. longipetiolata, KY609860, Phlegmariurus phlegmaria MT786212, Diphasiastrum digitatum MH549638, Lycopodium clavatum MH549642, Dendrolycopodium obscurum MH549637, Isoetes malinverniana MH549640, Isoetes yunguiensis MK047605, Selaginella moellendorffii HM173080, and Marchantia paleacea NC_001319.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The complete chloroplast genome sequence of Phlegmariurus phlegmaria in this study was submitted to the NCBI database under the accession number MT786212. https://www.ncbi.nlm.nih.gov/nuccore/?term=MT786212
Additional information
Funding
References
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Field AR, Testo W, Bostock PD, Holtum JAM, Waycott M. 2016. Molecular phylogenetics and the morphology of the Lycopodiaceae subfamily Huperzioideae supports three genera: Huperzia, Phlegmariurus and Phylloglossum. Mol Phylogenet Evol. 94(Pt B):635–657.
- Jin YW, Yang JB, Song Y, Yin TS, Li DZ. 2018. Get Organelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. bioRxiv. 256479.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Luo T, Li Y, Yuan X, Wang Y. 2019. The complete chloroplast genome sequence of Phlegmariurus carinatus. Mitochondrial DNA Part B. 4(2):3977–3978.
- Øllgaard B. 2015. Six new species and some nomenclatural changes in neotropical Lycopodiaceae. Nordic J Bot. 33(2):186–196.
- PPGI. 2016. A community-derived classification for extant lycophytes and ferns. J Syst E. 54:563–603.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wagner WH, Beitel JM. 1992. Generic classification of modern North American Lycopodiaceae. Ann Mo Bot Gard. 79(3):676–686.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.
- Wikström N, Kenrick P. 2000. Phylogeny of epiphytic Huperzia (Lycopodiaceae): paleotropical and neotropical clades corroborated by rbcL sequences. Nordic J Bot. 20(2):165–171.
- Zhang HR, Kang JS, Viane RLL, Zhang XC. 2017. The complete chloroplast genome sequence of Huperzia javanica (sw.) C.Y.Yang in Lycopodiaceae. Mitochondrial DNA B. 2(1):216–218.
