Abstract
Here, we present the complete mitochondrial DNA sequence of Dermacentor reticulatus. The mitogenome is 14,806 bp and contains 13 protein-coding, 2 rRNA, and 22 tRNA genes, along with 2 control regions. Dermacentor reticulatus mitogenome has the common mitochondrial gene order of Metastriata ticks. It is phylogenetically close to the mitogenomes of Dermacentor ticks, of which D. everestanus mitogenome is the closest with 85.7% similarity. These data provide insights into the phylogenetic relations among Dermacentor ticks.
The tick Dermacentor reticulatus (Fabricius, 1794) is widely distributed in Europe and Northern Asia and is the principal transmission vector for tick-borne infections, such as those caused by tick-borne encephalitis virus as well as Omsk hemorrhagic fever viruses, Rickettsia raoultii and Rickettsia slovaca (Wójcik-Fatla et al. Citation2011; Biernat et al. Citation2014; Karan et al. Citation2014; Shchuchinova et al. Citation2015; Zając et al. Citation2017; Chitimia-Dobler et al. Citation2019). Additionally, the genetic markers of Kemerovo and bluetongue viruses, Coxiella burneii, Bartonella quintana, and Theileria equi, and different species of Borrelia spp., Anaplasma spp, and Babesia spp have been detected in D. reticulatus. Recently, D. reticulatus has spread considerably and reached Siberian towns (Kartashov et al. Citation2019).
Here, we report the first complete mitochondrial DNA sequence of D. reticulatus. We collected adult ticks from a large town park in Tomsk (56°27’00.6“N 84°57'24.2“E) in 2017. Each tick was morphologically characterized and genetically identified with the sequencing fragment cox1 as previously described (Kartashov et al. Citation2019). The ticks were individually frozen in liquid nitrogen, crushed using plastic pestles, homogenized in phosphate-buffered saline, and treated with TRIzol reagent (Invitrogen Co., USA) to extract their DNA. Sequencing was performed using a MiSeq Reagent Kit v3 for 400 cycles. Raw reads and de novo contigs were aligned to D. everestianus mitogenome without inversions or transitions by using BWA (v.0.7.15). Of the 2409 aligned paired-reads, the mean length was 143 bp, and the coverage for the resulting mitogenome was 46,8. Gene annotation and sequence analysis were carried out through BLAST searches that queried the complete mitochondrial genome sequence of D. everestianus. ARWEN (v1.2) software was used to predict transfer RNA (tRNA) genes and their secondary structures and was adjusted to other frames (Jühling et al. Citation2012). For phylogenetic analysis, the mitochondrial sequences were derived from NCBI and aligned using MAFFT with the default settings (Katoh and Standley Citation2013).
The complete mitogenome of D. reticulatus was estimated at 14,806 bp, encoding 13 protein, 2 rRNA, and 22 tRNA genes and found to contain 2 control regions (MT478096). The mitogenome encoded 3609 amino acids in total. The A + T contents of the protein-coding genes ranged from 71.54% (cox1) to 80.49% (atp8). The order of the genes in D. reticulatus mitogenome and their transcriptional directions were found identical to those of D. marginatus, D. silvarum, and D. everestianus. The overall base composition of D. reticulatus mitogenome was determined to be 39.8% T, 12.5% C, 38.6% A, and 9.2% G. A + T ratio ranged from 74.0% (control region 1) to 89.4% (tRNA-Gly). D. reticulatus mitogenome was found 84.8%, 85.1%, 85.4%, and 85.7% identical to those of D. silvarum, D. marginatus, D. nutalii, and D. everestanus, respectively. The translation of 6 protein-coding genes (cox1, atp8, nd2, nd3, nd5, and nad6) was predicted to be initiated by the ATT start codon, of 1 gene (nad1) by the ATA codon, and 6 genes by the classical ATG codon. It was observed that most of the protein-coding genes terminated with TAA, whereas the translation of 4 genes was predicted to be stopped by the incomplete stop codon T. The 12S and 16S rRNA genes were estimated at 697 and 1204 bp, respectively.
Additionally, we observed that the 22 tRNA genes ranged between 56 bp (tRNA-Ser) and 69 (tRNA-Met) bp. Based on their sequences, we predicted these tRNAs to have the typical cloverleaf secondary structure, except for tRNA-Cys and tRNA-Ser1 (anticodon TCT), which lack the D-arms. Control regions 1 and 2 were found to be 311 and 305 bp, located between 12S rRNA and tRNA-Ile genes, and between tRNA-Leu and tRNA-Cys, respectively. We identified 94 noncoding nucleotides in 12 unassigned intergenic regions and short overlaps that totaled 46 bp at 5 gene junctions, with the largest overlap (26 bp) at the junction of tRNA-Glu and nd1. Phylogenetic analysis revealed that D. reticulatus and Dermacentor species clustered together with high statistical support, indicating that D. reticulatus belonged to the genus Dermacentor ().
Figure 1. Phylogenetic relationships between D. reticulatus and other tick species, based on mitochondrial sequences. The method of maximal likelihood with the GTR (G + I) algorithm was used (Guindon et al. Citation2010). Support indices were defined using the bootstrap method with 1000 repeats. MEGA X software was used for visualization (Kumar et al. Citation2018). The GenBank accession numbers and species of ticks are indicated on the tree. The genetic distance scale is shown at the bottom. The bold type was sequenced in this study.
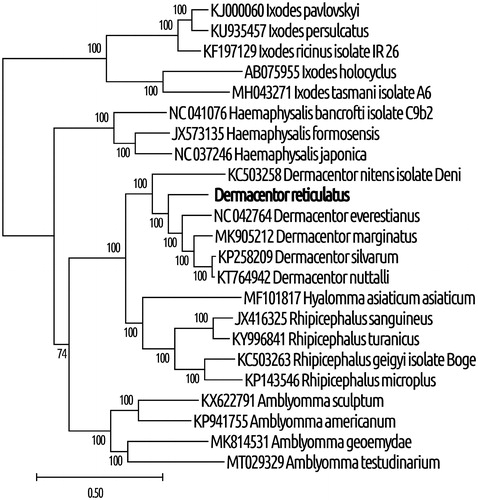
In conclusion, this study provides a new mitochondrion resource for phylogenetic studies and also novel genetic markers for further studies on Dermacentor ticks.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The collected ticks Dermacentor reticulatus (Fabricius, 1794) was archived in Collections of Tomsk State University (Tomsk, Russia) registered on “The Insect and Spider Collections of the World Website” under collection number UTR II 595.421.1 (http://hbs.bishopmuseum.org/codens/codens-inst.html).
The sequence data that support the findings of this study are openly available on GenBank using the accession number MT478096 (https://www.ncbi.nlm.nih.gov/nuccore/MT478096.1).
Additional information
Funding
References
- Biernat B, Karbowiak G, Werszko J, Stańczak J. 2014. Prevalence of tick-borne encephalitis virus (TBEV) RNA in Dermacentor reticulatus ticks from natural and urban environment, Poland. Exp Appl Acarol. 64(4):543–551.
- Chitimia-Dobler L, Lemhöfer G, Król N, Bestehorn M, Dobler G, Pfeffer M. 2019. Repeated isolation of tick-borne encephalitis virus from adult Dermacentor reticulatus ticks in an endemic area in Germany. Parasit Vectors. 12(1):90.
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Jühling F, Pütz J, Bernt M, Donath A, Middendorf M, Florentz C, Stadler PF. 2012. Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Res. 40(7):2833–2845.
- Karan LS, Ciccozzi M, Yakimenko VV, Lo Presti A, Cella E, Zehender G, Rezza G, Platonov AE. 2014. The deduced evolution history of Omsk hemorrhagic fever virus. J Med Virol. 86(7):1181–1187.
- Kartashov MY, Mikryukova TP, Krivosheina EI, Kuznetsov AI, Romanenko VN, Moskvitina NS, Ternovoi VA, Loktev VB. 2019. Genotyping of tick-borne infections in Dermacentor reticulatus ticks collected in urban foci of Tomsk. Parazytologiay. 53(5):355–369. (in Russian)
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Shchuchinova LD, Kozlova IV, Zlobin VI. 2015. Influence of altitude on tick-borne encephalitis infection risk in the natural foci of the Altai Republic, Southern Siberia. Ticks Tick Borne Dis. 6(3):322–329.
- Wójcik-Fatla A, Cisak E, Zając V, Zwoliński J, Dutkiewicz J. 2011. Prevalence of tick-borne encephalitis virus in Ixodes ricinus and Dermacentor reticulatus ticks collected from the Lublin region (eastern Poland). Ticks Tick Borne Dis. 2(1):16–19.
- Zając V, Wójcik-Fatla A, Sawczyn A, Cisak E, Sroka J, Kloc A, Zając Z, Buczek A, Dutkiewicz J, Bartosik K. 2017. Prevalence of infections and co-infections with 6 pathogens_in Dermacentor reticulatus ticks collected in eastern Poland. Ann Agric Environ Med. 24(1):26–32.
